'One stop shop'
In addition to enabling target assessment for drug discovery, canSAR has
also proved to be useful as a more general tool for the cancer research community. Rather than trawling through hundreds of individual journal articles and disparate datasets, canSAR provides a single portal to answer questions such as: What is known about this protein? In which cancers is it expressed or mutated? Are there any chemical tools that can be used to experimentally probe its activity? What drugs are approved and what clinical trials are underway?
In a 2016 article published in The Lancet Oncology, external scientists not involved in the work praised canSAR’s ‘one-stop shop approach to everything related to cancer drug discovery.’
Crucially, canSAR helps scientists to challenge their biases in drug discovery. There are many ‘hidden’ opportunities in the form of druggable proteins, but research shows that scientists generally focus on proteins and pathways that have already been well studied. canSAR allows scientists to be more objective based on the totality of the data and use this approach to generate new hypotheses and help prioritise which targets to pursue in drug discovery.
As the Lancet Oncology authors wrote: “It is often in these gaps of knowledge that the missing pieces of the drug-discovery puzzle are found.”
Stacking the cards
Here at the ICR, our scientists are using canSAR in studies across diverse cancer types, helping them to make the best decisions and prioritise the most promising drug targets.
Dr Adam Sharp is Leader of the Translational Therapeutics Team at the ICR and Honorary Consultant Medical Oncologist at The Royal Marsden NHS Foundation Trust. He has worked together with Professor Johann de Bono and the canSAR team to identify which potential drug targets to pursue related to prostate cancer.
Dr Sharp said: “It's about stacking the cards in your favour. We identify a lot of things that are interesting, but you want to know which ones may be more amenable for drug discovery before you commit the money and the time.”
Dr Sharp and colleagues identified a critical co-regulator of the androgen receptor, which is important to prostate cancer and can change as the cancer develops. Crucially it can mutate or lose part of its protein structure but remain active, meaning that the drugs used to target it can become ineffective over time.
Dr Sharp and colleagues found that a ‘co-chaperone’ protein, known as Bag-1L, seemed to stimulate the androgen receptor. Importantly, they found Bag-1L interacts with a region of the androgen receptor protein that is not currently targeted by existing drugs, and which because of features of its structure would be hard to drug.
However – as their recent study shows – targeting Bag-1L instead with future drugs may prove to be a fruitful way of tackling the androgen receptor by proxy, and could represent a new for patients with resistant prostate cancer.
Dr Sharp and Professor de Bono are now conducting further research on this protein and another target identified using canSAR called JMJD6 for possible future drugs.
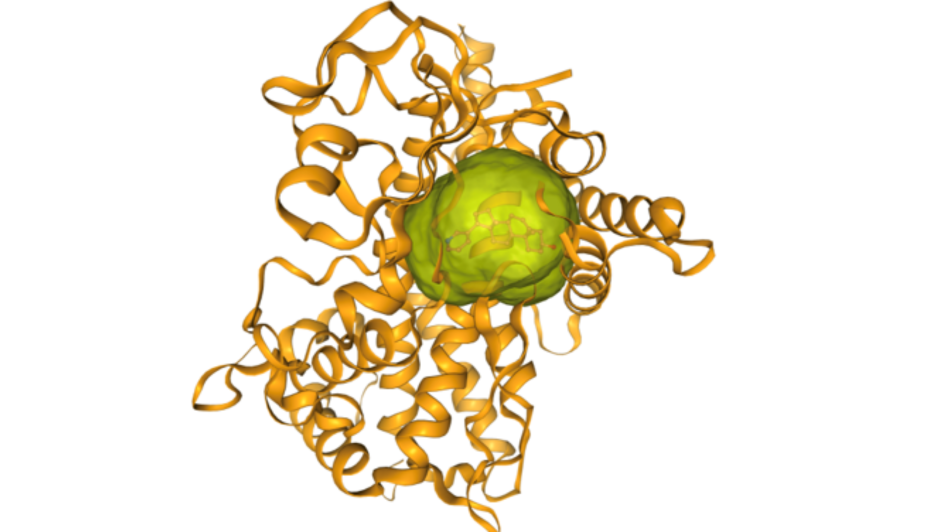
Image: Ligandable pocket found using canSAR in CYT P450 solved with abiraterone. Credit: Patrizio Di Micco
Powering and interfacing with other public resources
Currently, canSAR draws data from 21 different data source types and feeds into, and interfaces with, other resources. One of these is Probe Miner, developed at the ICR by Dr Albert Antolin, working with Professors Al-Lazikani and Workman.
To study and evaluate drug targets, chemical probes are incredibly valuable. These small-molecule compounds that can potently and selectively modulate the function of a particular protein of interest in cells or animals, allowing researchers to ask questions about their role and validate them as molecular targets. However, good chemical probes are hard to come by and biologists can find it hard to select the best chemical tools to use for their research.
Probe Miner helps by evaluating more than more than 1.8 million small molecules against more than 2,200 human targets and ranks them objectively against criteria that than can be pre-set or modified by the researcher. Importantly, Probe Miner is very complementary to another resource, the Chemical Probes Portal, led by our Chief Executive Professor Paul Workman, which is based on expert opinion. Use of both resources together is especially powerful.
For researchers who periodically or frequently use DepMap – the world’s largest cancer vulnerability screening initiative at the Broad Institute at Harvard and MIT – to mine genetic inactivation screens in human tumour cell lines to reveal cancer vulnerabilities, a built-in link means that users of that site can gain rapid and direct access to canSAR’s powerful druggability analysis of their target of interest with the click of a mouse.
Video: Celebrating 10 years of canSAR
Coronavirus canSAR
canSAR has always been a platform that thrives in finding connections in masses of complex and disconnected information.
When the Covid-19 pandemic hit, the canSAR team realised how vital the platform could be for researchers looking to rapidly repurpose drugs from across the whole of medicine, including cancer. It could be useful to find chemical probes and potential new targets. Importantly, despite the successful development of vaccines, finding drugs for patients affected by Covid-19 remains a high priority.
In 2020, recognising the large volume of data emerging on the coronavirus with little or no organisation of it, the ICR's canSAR team launched canSARS. The platform draws in data published across the world on viral proteins, interactions of viral proteins with human proteins, drugs and drug mechanisms, and clinical trials. It was the first portal of its kind for research on Covid-19 and related diseases such as SARS and MERS.
CanSARS was developed by the canSAR team funded by Cancer Research UK and Wellcome – and has already helped connect scientists across the globe with a wide range of expertise and disease interests who are working together to beat Covid-19.
It is especially important to help researchers cut through the noise and make objective assessments in the face of a torrent of data.
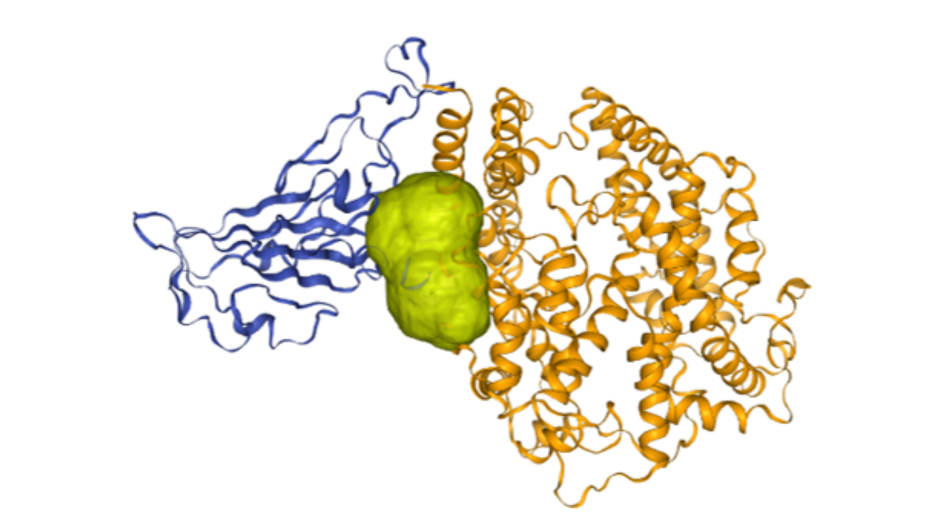
Image: Ligandable pocket found at the interface of Ace2-spike complex (Covid-19) using canSAR. Credit: Patrizio Di Micco
The future of canSAR
canSAR is a powerful integrated knowledgebase and decision support system for cancer drug discovery and translational cancer research. It helps to remove confirmation bias and guesswork by using big data and AI to objectively prioritise drug targets from hundreds of options. It also allows researchers to triage targets – by suggesting the next experiments to do to rule out inappropriate targets and to prioritise and fully validate the most promising ones for drug discovery.
To date, the canSAR team has published more than 50 research papers and the canSAR knowledgebase, data and approaches have been cited in more than 2,000 papers, with more emerging all the time as new findings are published and patents are filed. canSAR is free to use and the intellectual property rights remain with research teams who make the discoveries.
But the canSAR database is constantly evolving. There are no comparable platforms to canSAR for other diseases, and many researchers outside of oncology are already using the platform in unexpected ways – in addition to the coronavirus, canSAR has, for example, been cited in papers involved with Zika virus and psoriasis.
While canSAR’s impact is evident already, Professor Al-Lazikani says the best is yet to come: “canSAR’s first ten years of existence have no doubt saved decades of time in research and indeed uncovered many new drug targets that scientists may never have discovered or prioritised on their own.”
“Because canSAR helps at the beginning stages of drug discovery and this process can take decades, we could potentially see many new drugs thanks to canSAR in the future.”
Professor Paul Workman said: “As a researcher, I have always enjoyed working at the interfaces where different disciplines converge to enable unexpected discoveries. The next five to ten years will see incredible changes in how we use integrative big data analysis and AI to inform fundamental biological studies and drug discovery research for cancer and other diseases, enabled by large-scale scale public resources like canSAR.”

Image: canSAR team photo from 2019. The canSAR team is made up of chemists, structural biologists, bioinformaticians, software developers and computational biologists from all over the world. Photo credit: Helen Gunn