Abstract
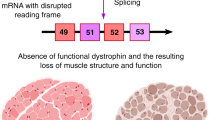
Frameshift mutations in the DMD gene, encoding dystrophin, cause Duchenne muscular dystrophy (DMD), leading to terminal muscle and heart failure in patients. Somatic gene editing by sequence-specific nucleases offers new options for restoring the DMD reading frame, resulting in expression of a shortened but largely functional dystrophin protein. Here, we validated this approach in a pig model of DMD lacking exon 52 of DMD (DMDΔ52), as well as in a corresponding patient-derived induced pluripotent stem cell model. In DMDΔ52 pigs1, intramuscular injection of adeno-associated viral vectors of serotype 9 carrying an intein-split Cas9 (ref. 2) and a pair of guide RNAs targeting sequences flanking exon 51 (AAV9-Cas9-gE51) induced expression of a shortened dystrophin (DMDΔ51–52) and improved skeletal muscle function. Moreover, systemic application of AAV9-Cas9-gE51 led to widespread dystrophin expression in muscle, including diaphragm and heart, prolonging survival and reducing arrhythmogenic vulnerability. Similarly, in induced pluripotent stem cell-derived myoblasts and cardiomyocytes of a patient lacking DMDΔ52, AAV6-Cas9-g51-mediated excision of exon 51 restored dystrophin expression and amelioreate skeletal myotube formation as well as abnormal cardiomyocyte Ca2+ handling and arrhythmogenic susceptibility. The ability of Cas9-mediated exon excision to improve DMD pathology in these translational models paves the way for new treatment approaches in patients with this devastating disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE database with the dataset identifier PXD014893. Unprocessed full scans of agarose gels and western blots for Figs. 1 and 3, Extended Data Figs. 1, 2 and 6–9 and Supplementary Figs. 3 and 5 are available online. Source data for Figs. 1–4, Extended Data Figs. 4–8 and Supplementary Figs. 2 and 3 are provided with the paper.
References
Klymiuk, N. et al. Dystrophin-deficient pigs provide new insights into the hierarchy of physiological derangements of dystrophic muscle. Hum. Mol. Genet. 22, 4368–4382 (2013).
Truong, D. J. et al. Development of an intein-mediated split-Cas9 system for gene therapy. Nucleic Acids Res. 43, 6450–6458 (2015).
Aartsma-Rus, A., Van Deutekom, J. C., Fokkema, I. F., Van Ommen, G. J. & Den Dunnen, J. T. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve 34, 135–144 (2006).
White, S. et al. Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization. Am. J. Hum. Genet. 71, 365–374 (2002).
Moser, H. Duchenne muscular dystrophy: pathogenetic aspects and genetic prevention. Hum. Genet. 66, 17–40 (1984).
Sharp, P. S., Bye-a-Jee, H. & Wells, D. J. Physiological characterization of muscle strength with variable levels of dystrophin restoration in mdx mice following local antisense therapy. Mol. Ther. 19, 165–171 (2011).
Van Deutekom, J. C. et al. Local dystrophin restoration with antisense oligonucleotide PRO051. New Engl. J. Med. 357, 2677–2686 (2007).
Goemans, N. M. et al. Systemic administration of PRO051 in Duchenne’s muscular dystrophy. New Engl. J. Med. 364, 1513–1522 (2011).
Verhaart, I. E. et al. The dynamics of compound, transcript, and protein effects after treatment with 2OMePS antisense oligonucleotides in mdx mice. Mol. Ther. Nucleic Acids 3, e148 (2014).
Bengtsson, N. E. et al. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat. Commun. 8, 14454 (2017).
EL Refaey, M. et al. In vivo genome editing restores dystrophin expression and cardiac function in dystrophic mice. Circ. Res. 121, 923–929 (2017).
Long, C. et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403 (2016).
Nelson, C. E. et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–407 (2016).
Tabebordbar, M. et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351, 407–411 (2016).
Amoasii, L. et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 362, 86–91 (2018).
Punnoose, A. R. et al. Cardiac disease burden and risk of mortality in hospitalized muscular dystrophy patients. Pediatr. Cardiol. 37, 1290–1296 (2016).
Feingold, B. et al. Management of cardiac involvement associated with neuromuscular diseases: a scientific statement from the American Heart Association. Circulation 136, e200–e231 (2017).
Vetter, A. et al. Adenoviral vectors coated with PAMAM dendrimer conjugates allow CAR independent virus uptake and targeting to the EGF receptor. Mol. Pharm. 10, 606–618 (2013).
Zincarelli, C., Soltys, S., Rengo, G. & Rabinowitz, J. E. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16, 1073–1080 (2008).
Pleger, S. T. et al. Cardiac AAV9-S100A1 gene therapy rescues post-ischemic heart failure in a preclinical large animal model. Sci. Transl. Med. 3, 92ra64 (2011).
Nelson, C. E. et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 25, 427–432 (2019).
Wagner, D. L. et al. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat. Med. 25, 242–248 (2018).
Chew, W. L. et al. A multifunctional AAV–CRISPR–Cas9 and its host response. Nat. Methods 13, 868–874 (2016).
Leyva-Leyva, M., Sandoval, A., Felix, R. & González-Ramírez, R. Biochemical and functional interplay between ion channels and the components of the dystrophin-associated glycoprotein complex. J. Membr. Biol. 251, 535–550 (2018).
Thajudeen, A. et al. Correlation of scar in cardiac MRI and high-resolution contact mapping of left ventricle in a chronic infarct model. Pacing Clin. Electrophysiol. 38, 663–674 (2015).
Fischer, C. et al. Long-term functional and structural preservation of precision-cut human myocardium under continuous electromechanical stimulation in vitro. Nat. Commun. 10, 117 (2019).
Moretti, A. et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. New Engl. J. Med. 363, 1397–1409 (2010).
Jiwlawat, N., Lynch, E., Jeffrey, J., Van Dyke, J. M. & Suzuki, M. Current progress and challenges for skeletal muscle differentiation from human pluripotent stem cells using transgene-free approaches. Stem Cells Int. 2018, 6241681 (2018).
Choi, I. Y. et al. Concordant but varied phenotypes among Duchenne muscular dystrophy patient-specific myoblasts derived using a human iPSC-based model. Cell Rep. 15, 2301–2312 (2016).
Young, C. S. et al. A single CRISPR–Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell. Stem. Cell. 18, 533–540 (2016).
Bar, S. et al. A novel product of the Duchenne muscular dystrophy gene which greatly differs from the known isoforms in its structure and tissue distribution. Biochem. J. 272, 557–560 (1990).
Kawaguchi, T. et al. Detection of dystrophin Dp71 in human skeletal muscle using an automated capillary western assay system. Int. J. Mol. Sci. 19, E1546 (2018).
Hinderer, C. et al. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an AAV vector expressing human SMN. Hum. Gene Ther. 29, 285–298 (2018).
Walter, M. C. & Reilich, P. Recent developments in Duchenne muscular dystrophy: facts and numbers. J. Cachexia Sarcopenia Muscle 8, 681–685 (2017).
Aartsma-Rus, A. et al. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum. Mutat. 30, 293–299 (2009).
US National Institutes of Health. Guide for the Care and Use of Laboratory Animals. NIH Publication No. 85–23 (NIH, 1996).
Richter, A. et al. Potential of primary kidney cells for somatic cell nuclear transfer mediated transgenesis in pig. BMC Biotechnol. 12, 84 (2012).
Kurome, M., Kessler, B., Wuensch, A., Nagashima, H. & Wolf, E. Nuclear transfer and transgenesis in the pig. Methods Mol. Biol. 1222, 37–59 (2015).
Concordet, J. P. & Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018).
Kupatt, C. et al. Cotransfection of vascular endothelial growth factor-A and platelet-derived growth factor-B via recombinant adeno-associated virus resolves chronic ischemic malperfusion: role of vessel maturation. J. Am. Coll. Cardiol. 56, 414–422 (2010).
Martin, P. & Bateson, P. Measuring Behaviour: An Introductory Guide (Cambridge Univ. Press, 2007).
Kupatt, C. et al. Endothelial nitric oxide synthase overexpression provides a functionally relevant angiogenic switch in hibernating pig myocardium. J. Am. Coll. Cardiol. 49, 1575–1584 (2007).
Hinkel, R. et al. Diabetes mellitus-induced microvascular destabilization in the myocardium. J. Am. Coll. Cardiol. 69, 131–143 (2017).
Pathik, B. et al. New Insights into an old arrhythmia: high-resolution mapping demonstrates conduction and substrate variability in right atrial macro–re-entrant tachycardia. JACC Clin. Electrophysiol. 3, 971–986 (2017).
Childers, M. K., Grange, R. W. & Kornegay, J. N. In vivo canine muscle function assay. J. Vis. Exp. 50, 2623 (2011).
Stauffer, W., Sheng, H. & Lim, H. N. EzColocalization: an ImageJ plugin for visualizing and measuring colocalization in cells and organisms. Sci. Rep. 8, 15764 (2018).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Perez-Riverol, Y. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019).
Gramlich, M. et al. Antisense-mediated exon skipping: a therapeutic strategy for titin-based dilated cardiomyopathy. EMBO Mol. Med. 7, 562–576 (2015).
Lian, X. et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 8, 162–175 (2013).
Dorn, T. et al. Interplay of cell–cell contacts and RhoA/MRTF-A signaling regulates cardiomyocyte identity. EMBO J. 37, e98133 (2018).
Fischer, B. et al. A complete workflow for the differentiation and the dissociation of hiPSC-derived cardiospheres. Stem Cell Res. 32, 65–72 (2018).
RStudio: Integrated Development for R. RStudio, Inc. (RStudio Team, 2015).
Labun, K. et al. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–w174 (2019).
Lindsay, H. et al. CrispRVariants charts the mutation spectrum of genome engineering experiments. Nat. Biotechnol. 34, 701–702 (2016).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–11.10.33 (2013).
Acknowledgements
We thank the patient with DMD and the healthy volunteer who provided blood for iPSC reprogramming. We acknowledge C. Scherb for technical assistance with the molecular cloning and G. Lederer (Cytogenetic Department, TUM) for karyotyping, as well as M. Ogris (MMCT Laboratory of Macromolecular Cancer Therapeutics, Department of Pharmaceutical Chemistry, University of Vienna, Austria) for advice on the G2 coating and A. Blutke (Research Unit Analytical Pathology, Helmholtz Centre Munich) for help with the pathological workup. A. Frank (Department of Apoptosis Research, Helmholtz Centre Munich) established the capillary western blot. M. Kösters (Gene Center, LMU) provided technical assistance with the mass spectrometry. The Muscular Dystrophy Association (USA) supports monoclonal antibody development in the laboratory of G. E. Morris, whom we thank for providing the MANDAG antibody against β-dystroglycan. This work was supported by grants from the Else Kröner-Fresenius Foundation (2015/180 and 2018/T20 to C.K., E.W. and W.W.), the European Research Council (ERC 788381 to A.M. and ERC 681524 to I.J.), German Research Foundation (DFG) Transregio Research Units 152 (to A.M. and K.L.L.), 127 (to E.W., A.S., A.B., N.K. and C.K.) and 267 (to A.M., A.S., K.L.L. and C.K.), and the German Center for Cardiovascular Research Munich Heart Alliance (to A.M., K.L.L. and C.K.). Generous support from A. Klesius and coworkers (Boston Scientific) with the Rhythmia system is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
C.K., E.W. and W.W. designed the pig study. M.K., V.Z., N.K., B.K. and E.W. generated the pigs with DMD and raised the cohort. L.F., A.B., K.K., R.H. and C.K. conducted the pig transduction, structural and functional analyses. P.H., C.J. and E.M. performed the high-resolution electrophysiological mapping and analyzed the data. T.B., K.K., R.H., I.J., K.V., V.J., F.A.R., S.R. and S. Krause performed and analyzed the expression assays and histology of pig tissues. F.G. and W.W. generated the intein-split Cas9 and gRNAs. H.B., A.G., S. Krebs, G.S. and F.G. sequenced and analyzed the DNA samples for the genome editing and off-target studies. T.B., T.Z. and A.W. generated and raised the AAV9 vectors. S.L. and T.Z. introduced the G2 optimization in vitro and in vivo. A.S. generated and analyzed the dTomato pigs for AAV-Cre transduction. A.M. and K.L.L. conceived of and supervised the iPSC study and provided financial support. A.B.M., D.S., T.H. and S.S. performed all of the experiments with iPSCs and their muscle derivatives. B.C. generated, characterized and differentiated the iPSC lines. A.B.M. generated the isogenic hDMDΔ51–52 hiPSCs. D.S., R.D. and T.D. analyzed the data. T.F. and F.F. performed the mass spectrometry. C.M.S., A.D. and D.S. performed the ex vivo experiments on heart slices and analyzed the data. S. Krause and M.C.W. provided human patient blood for reprogramming and conceptual advice. C.K. and A.M. wrote the paper. All authors commented on and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
C.K. and W.W. have filed a patent for G2-AAV9-Cas9-gE51. The other authors declare no competing interests.
Additional information
Peer review information Michael Basson was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 DMDΔ52 pig model and in-vitro testing of gene editing strategy.
a, Scheme of the generation and gene editing strategy of the DMDΔ52 pig model (left) and consequences of the genetic alterations at the protein level (right). DMD Exon 52 was replaced with a neomycin selection cassette (neoR), flanked by a murine PGK (muPGKprom) and an EM7 promoter (EM7prom), a bovine growth hormone polyadenylation signal (boGHpA) and loxP sites (lox). Blue arrows indicate splicike events. Asterisks indicate stop codons occurring in exon 53 due to reading frame incompatibility. Intended cutting sites for therapeutic gene editing are indicated by orange arrows. b, Scheme of gRNAs cloned into pAAV-N-Cas9 and pAAV-C-Cas9 vectors and tested in different combinations by transfection of porcine cells and PCR amplification (arrows: primer locations) of genomic DNA. c, Gel showing results of this test. d, Schematic representation of the intein-split-Cas9 system, consisting of two AAV constructs each harbouring one DMD-specific gRNA (gRNA DMD-5’ and gRNA DMD-3’, respectively) under the control of a U6 promoter and either the N-terminal or the C-terminal half of the Cas9 nuclease (N-Cas9(2−573) or C-Cas9(574−1368), respectively), fused to N- or C-terminal split-intein domains (N-intein and C-intein, respectively), under the control of a CBh promoter. NLS, nuclear localization sequence. FLAG, flag-tag. HA, HA-tag. e, Immunocytochemistry for both the N- and the C-terminal Cas9 peptides after AAV co-transduction of primary porcine myoblasts using a pair of AAV constructs (AAV9_5’_1 and AAV9_3’_1) harbouring gRNAs 5’-1 and 3’-1, respectively (representative for n=2). Scale bar, 15 µm. f, PCR analysis of genomic editing as in panel c in primary porcine kidney cells co-transduced with the above-mentioned pair of AAV constructs, with another pair (AAV9_5’_3 and AAV9_3’_3, harbouring gRNAs 5’-3 and 3’-3) or with control AAV, as indicated (representative for n=2 transductions).
Extended Data Fig. 2 Systematic analyses of DMD exon 51 deletion in pig skeletal muscles and heart after injection of AAV9-Cas9-gE51.
a,b, Top, genomic PCR analysis of DMD gene editing in samples from indicated skeletal muscles of DMD pigs treated by intramuscular (i.m) (a) or high dose intravenous (b) injection with G2-AAV9-Cas9-gE51, representative of 2 (a) and 3 (b) animals. Percentages of edited (ΔEx51+52) relative to total (ΔEx52 + ΔEx51+52) amplicon are shown. Quantifications by RT-PCR of the ratio of edited to total DMD mRNA expression (Δ51DMD / total DMD, middle) and mass spectrometry-based quantification of dystrophin protein (Dys) expression (bottom) are shown. C.l. = contralateral. c, Top, genomic PCR assessing cardiac DMD exon 51 editing in DMD pigs (five specimens of left ventricle (LV), left atrium (LA), right atrium (RA), right ventricle (RV), representative of 3 animals, are shown) treated with high dose intravenous injection of G2-AAV9-Cas9-gE51. Expected band sizes corresponding to unedited and edited DNA are indicated. Bottom, quantification of the ratio of edited to total DMD transcript (Δ51DMD/total DMD) by quantitative RT-PCR. d, Immunofluorescence for dystrophin (Dys) with wheat germ agglutinin (WGA) membrane staining in heart tissue of wildtype and untreated or high dose i.v. treated DMD pigs, representative of 8 images collected from 2 animals per group. Scale bars, 20 µm. e, left, immunoblotting for Cas9 in M. quadriceps muscle from 4 AAV9-Cas9-gE51 treated pigs as indicated, using an antibody against the Cas9 N-terminus. The expected band sizes corresponding to N-Cas9-N-intein (N-Cas9) and full-length Cas9 protein (Full Cas9) are indicated. Right, immunofluorescence staining of M. quadriceps muscle cells with antibodies detecting N-Cas9 (green) and the HA-tag (HA-C-Cas9) (yellow) with WGA membrane staining and DAPI nuclear labelling (DNA) (representative for n=2 pigs). Arrows indicate nuclei with overlapping fluorescence. Scale bar, 10 µm.
Extended Data Fig. 3 Analysis of off-target effects in porcine tissue samples by targeted deep sequencing.
For each off-target region, the reference sequence shows the gRNAs and PAM sequence marked by a black rectangle and additional 5 nucleotides up- and downstream. The tables show in the first line the number of sequence reads matching the reference sequence and in the following lines the number of INDELS found in each sample. Description of samples: Qc = quadriceps muscle; LV = left ventricle; Liv = liver; WT = wildtype, non-injected; i.m. = intramuscularly-injected; i.v. high = high dose intravenously-injected.
Extended Data Fig. 4 Colocalisation of dystrophin-associated glycoprotein complex (DGC) and restored dystrophin in DMD pig skeletal muscle after i.m. and i.v. injection of AAV9-Cas9-gE51.
a,b, Immunofluorescence co-staining for dystrophin and γ-sarcoglycan (a) or dystrophin and β-dystroglycan (b) in biceps femoris of wildtype, untreated DMD (DMD untr.) and intramuscularly (DMD i.m.) or intravenously (DMD i.v.) AAV9-Cas9-gE51-treated DMD pigs. Top and bottom rows for i.m. treated DMD in (a) are of areas close and distant to the injection site, respectively. Scale bars, 200 µm (left 20x merge column), 20 µm (right merge column), and 10 µm (detail column in b). c, Quantification of colocalization of dystrophin with either γ-sarcoglycan or β-dystroglycan. A threshold overlap score (TOS) was calculated giving a dimensionless number reflecting the degree of co-occurrence of signals between dystrophin and γ-sarcoglycan (TOS Dys-γSG, n=4 images except DMD i.v. n=6, collected from 2 pigs) or dystrophin and β-dystroglycan (TOS Dys-βDG, n=3 images from 2 pigs), with values ranging from 0 (no colocalisation) to 1 (perfect colocalisation). Data (Source Data Extended Data Fig. 4) are mean±SEM with p values from a one-way ANOVA with Bonferroni’s multiple comparison test (TOS Dys-γSG F=34.92, df=14; TOS Dys-βDG F=11.33, df=8).
Extended Data Fig. 5 In-vivo electro-mapping and ex-vivo single-cell Ca2+ analyses of DMD hearts.
a, Schematic drawing of the I8.5 F IntellaMap-Orion catheter used for high-resolution 3D-mapping, containing 64 flat microelectrodes (0.8 mm diameter) in a basket configuration with 8 splines that is steerable in 2 directions and can be opened and closed to provide appropriate wall contact for detection of electrophysiological signals. b, Mean voltage measured by in-vivo electro-mapping of the heart of wildtype (WT, n=3 animals), untreated DMD (n=2) and high dose intravenously (i.v.) G2-AAV9-Cas9-gE51 treated DMD (n=3) pigs (Source Data Extended Data Fig. 5), indicated as mean±SEM with p values from a one-way ANOVA with Tukey’s multiple comparison test (F=11.59, df=5). c, Size of endocardial low voltage area, expressed as percentage of the whole region, in indicated regions of the heart of WT (n=3 animals), untreated DMD (n=2) and high dose i.v. treated DMD (n=3) pigs (Source Data Extended Data Fig. 5), indicated as mean±SEM with p values from a two-way ANOVA with Tukey’s multiple comparison test (F=38.31, df=15). d, Schematic diagram of the experimental procedure for ex-vivo single-cell Ca2+ measurements, achieved by processing left-ventricular transmural sections to 1.0 x 0.5 cm myocardial tissue slices of 300 µm thickness, which were then submitted to physiological preload and continuous electrical field stimulation in biomimetic culture chambers. The right panel shows a pseudocolor image of Fluo-4 fluorescence recorded from a slice loaded with this calcium sensor and a region of interest (ROI) over which the average fluorescence signal was calculated to investigate intracellular calcium dynamics.
Extended Data Fig. 6 Generation of patient-specific DMD iPSC isogenic lines.
a, Left, schematic representation of the DMD exon 52 deletion in the patient-specific hDMDΔ52 hiPSCs and position of the primers (DMD exon 52 fwd and DMD exon 52 rev) used for PCR verification of the mutation. Right, a gel showing the 370 bp amplicon specific for the exon 52 deletion. Bottom, results from Sanger sequencing of the hDMDΔ52 hiPSCs. b, Bright field image of alkaline phosphatase staining in hDMDΔ52 hiPSC colonies at passage 6. Scale bar, 100 µm. c, Normal karyotype in hDMDΔ52 hiPSCs at passage 23. d, RT-PCR analysis of the Sendai vector (SeV) and transgenes OCT4, SOX2, KLF4 and c-MYC in untransduced peripheral blood mononuclear cells (PBMCs, negative control), Sendai-transduced PBMCs (positive control) and hDMDΔ52 hiPSCs at passage 13, using GAPDH as an endogenous control. e, Immunofluorescence analysis of the pluripotency markers NANOG and TRA-1-81 in hDMDΔ52 hiPSCs at passage 24. Scale bar, 50 µm. f, RT-qPCR analysis of the pluripotency markers OCT4, SOX2, NANOG, REX1 and TDGF-1 in hDMDΔ52 hiPSCs. The relative mean fold change expression normalized to GAPDH is indicated, n=2 (passages 13 and 20). g, RT-qPCR analysis of markers of endoderm (SOX7, AFP), mesoderm (CD31, DES, ACTA2, SCL, CDH5) and ectoderm (KRT14, NCAM1, TH, GABRR2) after 21 days of spontaneous embryoid body differentiation of hDMDΔ52 hiPSCs. The relative mean fold change expression normalized to GAPDH is indicated, n=2 independent differentiations. h, Left, schematic diagram of the deletion of DMD exon 51 in hDMDΔ52 hiPSCs and primers used for PCR verification of the deletion (right). i, Normal karyotype after CRISPR/Cas9 editing confirmed in hDMDΔ51-52 hiPSCs at passage 14. Uncropped gels for (a), (d), and (h) and statistics for (f) and (g) are shown in Source Data Extended Data Fig. 6.
Extended Data Fig. 7 Generation of control iPSCs from a healthy, young male donor.
a, Bright field image of alkaline phosphatase staining performed on control hiPSC colonies at passage 12. Scale bar, 100 µm. b, Normal male karyotype confirmed in control hiPSCs at passage 21. c, RT-PCR analysis of the Sendai vector (SeV) and transgenes OCT4, SOX2, KLF4 and c-MYC in untransduced peripheral blood mononuclear cells (PBMCs, negative control), Sendai-transduced PBMCs (positive control) and control hiPSCs at passage 24, using GAPDH as an endogenous control. d, Immunofluorescence analysis of the pluripotency markers NANOG and TRA-1-81 in hiPSCs at passage 21. Scale bar, 50 µm. e, RT-qPCR analysis of the pluripotency markers OCT4, SOX2, NANOG, REX1 and TDGF-1 in hiPSCs. The mean fold change expression relative to parental patient PBMCs and normalized to GAPDH is indicated, n=2 (passages 15 and 21). f, RT-qPCR analysis of markers of endoderm (SOX7, AFP), mesoderm (CD31, DES, ACTA2, SCL, CDH5) and ectoderm (KRT14, NCAM1, TH, GABRR2) in control hiPSCs after 21 days of spontaneous embryoid body differentiation. The mean fold change expression relative to hiPSCs and normalized to GAPDH is indicated, n=2 independent differentiations. Uncropped gels for (c) and statistics for (e) and (f) are shown in Source Data Extended Data Fig. 7.
Extended Data Fig. 8 Direct infection of hDMDΔ52 hiPSC-derived skeletal myoblasts and cardiomyocytes with AAV6-Cas9-gE51 restores expression of a re-framed dystrophin.
a, Bright field images of skeletal myoblasts from control, hDMDΔ52 or hDMDΔ51-52 hiPSCs, representative of >10 images (3 independent differentiations). Scale bars, 100 µm. b, RT-qPCR analysis of MYOD1, MYOG and DES in control (n=5 independent differentiations except DES n=4), untreated hDMDΔ52 (n=4 except MYOD1 n=3), hDMDΔ52 6 days after AAV6-Cas9/gE51 transduction (hDMDΔ52+AAV, n=3) or hDMDΔ51-52 (n=2) myoblasts. Relative fold change expression normalized to GAPDH is shown as mean±SEM with p values from a one-way ANOVA with Bonferroni’s multiple comparison test (MYOD1 F=12.86, df=10; MYOG F=7.159, df=9; DES F=103, df=9). c, Images of hDMDΔ52 skeletal myoblasts (top) or cardiomyocytes (bottom) 10 days after transduction with AAV6-Cas9/gE51 vectors encoding eGFP or mCherry (AAV6-N-Cas9/gRNA5’-eGFP and AAV6-C-Cas9/gRNA3’-mCherry), representative of >20 images (2 independent differentiations). Scale bars, 100 µm. d, Percentages of double-positive, single-positive and double-negative skeletal myoblasts (top) or cardiomyocytes (bottom) 4 and 10 days after transduction. e, Genomic PCR analysis of DMD exon 51 excision after AAV6-Cas9/gE51 transduction of hiPSC-derived skeletal myoblasts or cardiomyocytes (3 independent differentiations). f, Percentage of exon 51 excision based on relative PCR band intensity (edited versus total), indicated as mean±SEM. g, Dystrophin detection by capillary-based immunoassay after myotube induction of control, untreated or AAV6-Cas9/gE51-transduced hDMDΔ52 and hDMDΔ51-52 skeletal myoblasts (top) and in control, untreated or AAV6-Cas9/gE51-transduced hDMDΔ52 and hDMDΔ51-52 cardiomyocytes (bottom) from 3 independent differentiations. Bands represent the main (Dp427) and a shorter dystrophin isoform (Dp71). β-actin, loading control. h, Dystrophin (Dp427) levels normalized to β-actin expressed as percentage of mean level in control cells are depicted for skeletal muscle cells (top) and cardiomyocytes (bottom) as mean±SEM (p values from one-way ANOVA with Bonferroni’s multiple comparison test; Skeletal cells F=63.46, df=8, Cardiomyocytes F=21.59, df=8).
Extended Data Fig. 9 AAV6-Cas9/scrambled-gRNA transduction of hDMDΔ52-iPSC-derived myoblasts fails to restore dystrophin expression and capability of the cells to differentiate into myotubes.
a, Immunofluorescence staining for myosin heavy chain β (MyHC-β), α-actinin and dystrophin 14 days after skeletal myotube induction of untreated hDMDΔ52 myoblasts, hDMDΔ52 myoblasts transduced with AAV6-Cas9/gE51 (hDMDΔ52 + AAV) or hDMDΔ52 myoblasts transduced with AAV6-Cas9/scrambled-gRNA (hDMDΔ52 + AAV-scr) (representative for n=3 independent differentiations). Scale bars, 100 µm. b, Genomic PCR analysis of DMD exon 51 excision 14 days after skeletal myotube induction of untreated hDMDΔ52 myoblasts, hDMDΔ52 myoblasts transduced with AAV6-Cas9/gE51 (hDMDΔ52 + AAV) or hDMDΔ52 myoblasts transduced with AAV6-Cas9/scrambled-gRNA (hDMDΔ52 + AAV-scr), representative of 2 independent differentiations. The expected band sizes corresponding to edited and unedited genomic DNA are indicated. Uncropped gel is shown in Source Data Extended Data Fig. 9. c, Capillary-based immunoassay of dystrophin 14 days after skeletal myotube induction of untreated hDMDΔ52 myoblasts and hDMDΔ52 myoblasts transduced with AAV6-Cas9/gE51 (hDMDΔ52 + AAV) or AAV6-Cas9/scrambled-gRNA (hDMDΔ52 + AAV-scr), using β-actin as a loading control, representative of 2 independent differentiations. The antibody detected both the main dystrophin isoform (Dp427) and a shorter isoform (Dp71). Uncropped blots are shown in Source Data Extended Data Fig. 9.
Extended Data Fig. 10 Levenshtein analysis of distance of guide RNAs around variants identified by whole-genome sequencing of isogenic hDMDΔ51-52 iPSCs compared to the parental hDMDΔ52 iPSCs.
Histogram of all minimal Levenshtein distances obtained by aligning the two DMD-E51 guide RNAs to all variants identified by whole-genome sequencing in isogenic hDMDΔ51-52 iPSCs compared to the parental hDMDΔ52 iPSC line, applying a sliding window starting 25 bp upstream and ending 25 bp downstream of each variant. For any candidate region around a variant at least 7 operations (base exchanges, deletions, insertions) were required to match one of the gRNAs, indicating that the variants were not off-target effects of the CRISPR-Cas treatment.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5, Tables 1–7 and source data for Supplementary Figs. 2, 3 and 5.
Supplementary Video 1
Footage of a camera observing the animal cage and showing the sudden death of one of the two DMDΔ52 pigs (animals with a blue mark on the back).
Supplementary Video 2
Spontaneous contracting skeletal myotubes from healthy control hiPSCs, representative of three independent differentiations.
Supplementary Video 3
Spontaneously contracting skeletal myotubes from patient hDMDΔ52 hiPSCs obtained after AAV6-Cas9-gE51 transduction of hDMDΔ52 hiPSC-derived myoblasts, representative of three independent differentiations.
Supplementary Video 4
Spontaneously contracting skeletal myotubes from isogenic patient hDMDΔ51–52 hiPSCs, representative of three independent differentiations.
Source data
Source Data Fig. 1
Statistical source data
Source Data Fig. 1
Unprocessed gels/western blot
Source Data Fig. 2
Statistical source data
Source Data Fig. 3
Statistical source data
Source Data Fig. 3
Unprocessed western blot
Source Data Fig. 4
Statistical source data
Source Data Extended Data Fig. 1
Unprocessed gels
Source Data Extended Data Fig. 2
Unprocessed gels, blots
Source Data Extended Data Fig. 4
Statistical source data
Source Data Extended Data Fig. 5
Statistical source data
Source Data Extended Data Fig. 6
Statistical source data
Source Data Extended Data Fig. 6
Unprocessed gels
Source Data Extended Data Fig. 7
Statistical source data
Source Data Extended Data Fig. 7
Unprocessed gels/capillary western blot
Source Data Extended Data Fig. 8
Statistical source data
Source Data Extended Data Fig. 8
Unprocessed gels/capillary western blot
Source Data Extended Data Fig. 9
Unprocessed gels/capillary western blot
Rights and permissions
About this article
Cite this article
Moretti, A., Fonteyne, L., Giesert, F. et al. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat Med 26, 207–214 (2020). https://doi.org/10.1038/s41591-019-0738-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0738-2
This article is cited by
-
Präzisionsmedizin vertieft die personalisierte Medizin in der Kardiologie
Die Innere Medizin (2024)
-
Recent advances in CRISPR-based genome editing technology and its applications in cardiovascular research
Military Medical Research (2023)
-
Sarcomeric network analysis of ex vivo cultivated human atrial appendage tissue using super-resolution microscopy
Scientific Reports (2023)
-
Cas9-mediated replacement of expanded CAG repeats in a pig model of Huntington’s disease
Nature Biomedical Engineering (2023)
-
Forced activation of dystrophin transcription by CRISPR/dCas9 reduced arrhythmia susceptibility via restoring membrane Nav1.5 distribution
Gene Therapy (2023)