Abstract
Nanoparticles promise to improve the treatment of cancer through their increasingly sophisticated functionalisations and ability to accumulate in certain tumours. Yet recent work has shown that many nanomedicines fail during clinical trial. One issue is the lack of understanding of how nanoparticle designs impact their ability to overcome transport barriers in the body, including their circulation in the blood stream, extravasation into tumours, transport through tumour tissue, internalisation in the targeted cells, and release of their active cargo. Increased computational power, as well as improved multi-scale simulations of tumours, nanoparticles, and the biological transport barriers that affect them, now allow us to investigate the influence of a range of designs in biologically relevant scenarios. This presents a new opportunity for high-throughput, systematic, and integrated design pipelines powered by data and machine learning. With this paper, we review latest results in multi-scale simulations of nanoparticle transport barriers, as well as available software packages, with the aim of focussing the wider research community in building a common computational framework that can overcome some of the current obstacles facing efficient nanoparticle design.
Similar content being viewed by others
Introduction
For over three decades, nanomedicine has held the potential to prevent, delay, control, or even cure cancer. Over two-thirds of nanomedical research has been focused on oncology1,2,3. Starting with Doxil in 1995, there are now around a dozen FDA-approved anti-cancer nanoparticles4. Nanoparticles, typically between 1 and 100 nm in size5, have been shown to be effective drug-vectors thanks to their ability to shield therapeutic cargos throughout transport, accumulate in certain tumour types6, activate through environmental (e.g. pH, enzymatic activity) or external stimuli (such as magnetic resonance, ultrasound, or infrared light7,8,9), or target specific cells10. Active targeting includes designs where the nanoparticle surface is functionalized to bind to target receptors or other proteins expressed on the membrane of the cancer cells, the extracellular matrix (ECM), or tumour vasculature. On the other hand, passive targeting is mediated by the size of nanoparticles, which is thought to favour their accumulation at certain tumour sites due to the EPR effect6. Increasingly sophisticated nanoparticle functionalisation, or combination strategies, are being explored as mechanisms to make treatments both smarter and more effective11.
The versatility of nanomedicine is a result of the vastness of the design space for nanoparticles. Changing the behaviour of nanoparticles can be achieved by altering the conventional 4S parameters, namely size, shape, surface functionalisation and stiffness12. The surface functionalisation in particular contributes to the charge of the particle, its stability in (and clearance from) the bloodstream, as well as active targeting. Further specifications include the material used (for example, energy receptive or reactive to the environment), and the loading of the drug (such as chemotherapy).
Key to predicting clinical impact, such as the correct distribution of nanoparticles over a tumour, relies on an accurate understanding of the interaction between individual nanoparticles and their environment. These interactions can motivate coordinated strategies, where sub-populations of nanoparticles might alter their local environment (such as degrading the ECM13), amplify signals to other nanoparticles14, create anchored binding sites for other nanoparticles, or self-assemble/disassemble to improve transport15,16. Such coordinated strategies, sometimes inspired by swarm intelligence11, demonstrate the high degree of customisability inherent to nanoparticle design. While this customisability can lead to novel nanoparticle strategies, their discovery and validation can be a challenge.
Despite, or perhaps because of, the many design parameters that can alter the functioning of nanoparticles, many prototypes fail to realise success as an alternative (or supplement) to conventional treatment techniques. Though many pre-clinical trials demonstrate increased specificity, only 5% of the dose typically reaches the tumour site during clinical trials17. Transport barriers to the target site are expected to play a key role in this disappointing result, including travel through the circulatory system and avoiding clearance, extravasation to the tumour site, tissue penetration within the tumour, and delivery to the relevant part of the cell.
Recent advances in computational power, as well as improved simulations of nanoparticles, and the biological transport barriers that affect them, allow for multi-scale simulations that can investigate the influence of a range of parameters in biologically realistic scenarios. This presents a new opportunity for high-throughput, systematic, and integrated nanoparticle-design pipelines. Such future pipelines may enable general design principles which, when combined with patient-specific data, could provide personalised treatment and care. Furthermore, utilisation of in silico models can minimise the costs associated with more conventional trial and error approaches in the laboratory, especially when combined with recent machine learning techniques such as ‘active learning’18.
There are now many computational models and results that have been reported on different stages of tumour initiation, growth, and the interaction of nanoparticles within the body and tumour. Below, we summarise some of the most recent and relevant in silico models for overcoming nanoparticle transport barriers. We focus predominantly on cancer nanomedicine, but note that many of the models we describe are relevant to other cancer therapies that may not use nanoparticles, such as immunotherapy or chemotherapy, as highlighted in several reviews19,20,21.
Many of the in silico models that we describe have been integrated with in vitro and in vivo experiments, as well as with machine learning techniques. We believe that a systematic and integrated framework for drug development, building on these examples, can minimise costly trial-and-error approaches and accelerate the development of effective nanoparticle cancer therapies. However, such a framework requires a shared modelling language and a multidisciplinary approach, with ongoing collaboration between mathematical and computational modellers, experimentalists, and clinicians. This paper is an attempt to provide a comprehensive review that sets the stage for such a pipeline, as we detail in our concluding remarks.
In silico models of tumours
Central to in silico modelling of nanoparticle treatments is having realistic models of the tumours. Malignant tumours caused by the uncontrolled proliferation of faulty cells, can be separated into three groups; carcinomas, leukomas, and sarcomas. Of the three groups, carcinomas are the most common (accounting for 90% of reported cases22) and predominantly appear in certain cell types such as breast, lung, prostrate, and colon/rectum. These cell types alone accounted for more than half of all cancers reported in the USA in 201823. Carcinomas are typically large, greater than 1 cm3 when detected, and fatal if left untreated22. They are the main topic of this paper.
Existing mathematical approaches24 can be separated into three groups: continuum, discrete, and hybrid. The continuum models, which utilise ordinary and partial differential equations, are typically faster to compute and are better suited to capturing global changes to a tumour, such as availability of oxygen and nutrients (e.g. to predict the development of a hypoxic core in a solid tumour). However, they are limited in their ability to recreate heterogeneity, single-cell interactions and other features better suited to a discrete modelling approach such as agent-based (AB), cellular potts (CP), and cellular automata (CA). Discrete models focus on the individual units (such as the cell) that follow a simple set of rules guiding their growth, death, and interaction with the local environment and other agents. This could be used to model the realistic heterogeneous development of a tumour over time, how cellular resistance emergence, or the growth of angiogenic vessels.
Both approaches have their strengths and weaknesses, and this has led some researchers to combine both continuum and discrete approaches to gain the benefits of both, so-called hybrid methods. Hybrid models build on discrete models but combine them with gradients of variables, modelled using continuum equations25. These models are also able to monitor individual cell behaviour such as mutation as well as the influence of local environmental variables. For a review of discrete and hybrid models and their application to cancer, see, for example work by Kim et al. and An et al.26,27, for multi-scale cancer modelling, see work by Deisboeck et al. and Norton et al.28,29, and for agent-based cancer models, see work by Metzcar et al.30.
Modelling transport barriers for nanoparticle delivery
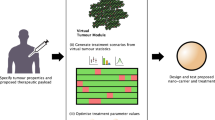
The use of nanoparticles as drug-delivery vectors requires that the engineered particles navigate from point of entry into the body to their prescribed biological target. The nanomedicine is normally administered topically, orally, intravenously or through direct injection to the site (e.g. to overcome the blood-brain barrier). Transportation barriers will present themselves immediately. Effective use of NPs requires overcoming these transport barriers: travel through the vasculature31,32,33, extravasation34,35,36, avoiding uptake by the reticulo-endothelial system (RES)37,38,39, progression through the tumour tissue35,40,41,42,43,44, endocytosis45,46,47,48,49,50, and delivery to the relevant part of the cell51,52,53. While each of these barriers presents a considerable individual challenge, the optimum NP design must seek to overcome several (if not all) of them to reach maximum efficacy. However, the high number of parameters that can be altered when designing nanoparticles results in a rich design space which is difficult to optimise, as shown in Fig. 1.
Circulation, clearance (CC) and extravasation (EV)
For intravenously injected nanoparticles, remaining in the blood stream as long as possible (long half-life) will favour their accumulation in certain types of tumours that benefit from the leaky nature of angiogenic blood vessels. The longer the circulation time, the higher the probability of escaping into the tumour site through multiple arrival ports (as opposed to a single one with direct injection54). Several direct factors influence circulation time including nanoparticle size, shape, and charge. Nanoparticles that are smaller than 5 nm are quickly cleared by the kidneys55. Nanoparticles larger than 100 nm are more likely to be detected by macrophages and cleared56. Between 5 and 100 nm, the charge of nanoparticles and resulting protein corona, or opsonisation, will drive macrophage uptake57. In silico methods such as molecular dynamics simulations have been used to examine the influence of these factors (such as protein corona formation and pH-stability) on nanoparticle transport58,59,60. For a further review of factors that affect circulation time for nanoparticles, see, for example, work by Yoo et al.61.
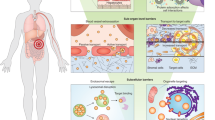
There are other influencing factors that can affect nanoparticle transport through the vascular system, as shown in Fig. 2. For example, margination occurs when red blood cells segregate in the centre of the vessel, creating a ‘cell-free’ layer near to the vessel wall57. This allows for nanoparticles to more readily escape from the porous tumour vessel by avoiding interactions with the blood cells and was demonstrated in silico, using a combination of dissipative particle dynamics (DPD) (a coarse-graining approach) and computational fluid dynamics62. By modelling the interaction between nanoparticles, red blood cells and white blood cells in the vessel, it was shown that the larger nanoparticles (greater than 500 nm) are able to take advantage of this margination effect whereas smaller nanoparticles (less than 200 nm) are trapped with the red blood cells in the ‘core’ region12.
Alternatively, the clustering of red blood cells may prove an important factor in increasing the biodistribution of nanoparticles, as clustering behaviour has been observed to create small variations in the capillary radius that can lead to drastic changes in the blood flow57. An agent-based model was also used to investigate the influence of red blood cells on blood-mediated nanoparticle62. It was shown that polydispersity of nanoparticles is an important factor, which can influence clearance, tissue penetration, and the immune system interaction.
Physiologically based pharmacokinetic models are particularly well suited to predicting nanoparticle transport through the body, where each relevant region is reduced into compartments and where each compartment contains permeability-limited, or perfusion-limited, models that describe how drugs, molecules, or indeed nanoparticles, pass through the individual compartments. The models are parameterised by properties that can be measured in vitro or in vivo, such as tissue volume or tissue flow rate, while drug-specific properties, such as clearance and tissue partition coefficients, are either scaled based on in vitro measurements or found using other in silico models where in vitro measurements are challenging to obtain. These pharmacokinetic models have been applied to nanoparticle-based drug delivery therapies, predicting, for example, the plasma and tissue concentrations of nanoparticles in mice63. For a review of pharmacokinetic models, see, for example, work by Yuan et al.64.
Along with transport through the body, it is important to be able to characterise the cytotoxicity of nanoparticle-based treatments40,63,65. This is because nanoparticles are often used to address high levels of toxicity caused by other anti-cancer treatments such as chemotherapies, can be given in conjunction with such treatments, and are often administered to immunocompromised patients. Computational models have been developed to explore the cytotoxicity of these therapies. For example, pharmacokinetic models have been used to investigate the difference in interspecies cytotoxicity, an important factor in translational research63. Alternatively, a continuum model of tumour growth was combined with experimental work to evaluate the cytotoxicity of gold nanoparticles carrying chemotherapy molecules40. It was shown that nanoparticle-based therapies had an overall increase in efficacy and reduced cytotoxicity when compared to the free drug. For a review of the application of computer models to the cytotoxicity of nanoparticles, see, for example, work by Ding et al.65.
One of the many benefits of nanoparticle-based drug delivery is that nanoparticles can be designed to take advantage of the so-called enhanced permeability and retention (EPR) effect which allows the nanoparticles to passively target the tumour. EPR is a result of the leaky angiogenic micro-vessels that cause an increase in the fluid pressure within the tumour66. Nanoparticles tend to escape from the enlarged pores in the faulty tumour vascular network into the surrounding tissues. In silico investigations have highlighted potential best nanoparticle-design to take advantage of irregularities at the endothelial wall. For example, the influence of increasing vascular permeability has been considered and shown to be beneficial for nanoparticle delivery36. However, the EPR effect has substantial variability both temporally and spatially and cannot be assumed for all tumours (it is not found in gastric and pancreatic cancer, for example work by Nakamura et al.67). Furthermore, the high interstitial pressure developed in certain tumours can also limit the in-flow of nanoparticles from outside the tumour68 which can discount the therapeutic usefulness of the EPR effect in humans. Hence, care should be given when assuming this means of passive targeting68.
One potential cause for this discrepancy in the usefulness of the EPR effect may lay with the animal models that are used to study it. For example, the rapid angiogenesis that is induced for in vivo experiments leads to tumours that grow on much faster timescales than commonly occurring in humans. Furthermore, the ratio of grown animal tumour to the total weight of the animal used will be far higher than that observed in humans. Both these features will promote the extravasation potential of nanoparticles, potentially to unrealistic levels when compared against clinical studies38,69. As discussed above, the EPR effect can be strong in some tumours but negligible in others and should not be assumed as a general feature for all tumour types68.
The design of the nanoparticles can have implications both for the success of the individual nanoparticles as well as subsequent injections. For example, if large particles are injected first then their reduced diffusive ability can clog extravasation points for successful transport of smaller nanoparticles. This indicates a possible two-phase extravasation profile where fast extravasation is followed by slow extravasation, highlighting additional design considerations when engineering nanoparticles69. The structure of the vascular network will also be influential, especially around the tumour site. For example, spatial irregularities within the network can lead to uneven distribution of drugs. This has been considered in work by Sefidgar et al.32 where the concentration of drug within the interstitial flow was investigated using a multi-scale model of a heterogeneous and dynamic vascular network around a tumour. By allowing feedbacks between hemodynamic and metabolic stimuli and the capillary network, a more irregular capillary network was generated with high interstitial fluid pressure (IFP) within the tumour, as typically observed in vivo. It was shown that the elevated IFP within this irregular network led to a heterogeneous distribution of drugs around the tumour region, highlighting that static or regular vascular networks may be an overly restrictive assumption when modelling drug delivery.
As extravasation is central to nanoparticle penetration and accumulation, it is important that general design principles are known to optimally extravasate into the tumour. Again, in silico models are of use in testing various designs. For example, Brownian dynamics were used to investigate the influence of nanoparticle diameter and aspect ratio on extravasation and demonstrated that larger aspect ratio increases extravasation by alignment with the streamlines exiting the pore34. Alternatively, models that describe nanoparticle vascular transport also include considerations of extravasation to improve model accuracy36,70. Both vascular transport and extravasation at the tumour site can be modelled using agent-based models33. This has been used to simulate tumour growth with a realistic vessel network structure (when compared against 3D intravital image). Though this has yet to be coupled with a nanoparticle design framework, this may offer a further tool for bioengineers to trial designs prior to in vitro and/or vivo testing.
Tissue penetration (TP)
For nanoparticle-based therapies, a substantial challenge is predicting the depth that the NPs are able to penetrate into the tumour and where they accumulate, as shown in Fig. 3. In healthy tissue, the IFP leads to the necessary pressure gradient to transport nanoparticles away from vessels. However, IFP within tumour environments can lead to additional barriers for drug delivery as well as driving the growth of the tumour. Experiments in vivo have shown that IFP is uniform across much of the tumour but drops significantly at the edge71. This uniform pressure creates a diffusive environment across much of a tumour and a steep outward flow around the edge. The diffusive environment within the tumour means that extravasated particles move slowly and can diffuse back into the capillary system rather than entering deeper into the tumour tissue35,72. Near the edge of a tumour, there is a risk that the nanoparticles will not be retained, instead being irreversibly pushed by the steep pressure gradient37.
NPs (shown as black dots) are shown leaving the vascular system and diffusing through towards endothelial cells. NPs are required to penetrate deep enough through the tumour tissue in order to be effective while aiming to be internalised by specific cell types (cancer cells and cancer stem cells) while avoiding other cell types (such as healthy cells).
In silico models offer a means of understanding the obstacles to tumour penetration. The influence of the IFP on the blood and lymphatic systems and their influence on drug-delivery has been investigated using a combination of fluid dynamics and agent-based modelling73. This work demonstrated how drug distribution increases as the lymphatic response decreases, due to a reduction in the clearance of drug-delivery vectors. It also demonstrated that the dual normalising of both the vasculature and interstitial is required to improve drug efficacy in order to simultaneously reduce the IFP within the tumour as well as minimise the heterogeneity of the drug distribution within the tumour tissue.
The heterogeneous distribution of nanoparticles has recently been considered using in silico models that combines the growth of a tumour with angiogenesis and drug delivery74. This work used a three-dimensional model continuum approach to highlight conditions of the tumour for enhanced nanoparticle drug delivery such as high interstitial porosity (to enhance nanoparticle transport). Alternatively, the connection between tumour growth and avascular network generation was modelled using a multi-scale approach that coupled tumour growth with nanoparticle transport75. The binding affinity of nanoparticles was shown to play a role in the accumulation within the tumour, where those with high binding affinity concentrate near the vessels and that slow nanoparticles failed to penetrate at lethal levels throughout the tumour tissue76.
This coupled model of tumour growth, vasculature system and nanoparticle adhesion has since been used as a tool for optimising nanoparticle design77. Here, the model was used to find the optimal nanoparticle diameter for accumulation and penetration. Alternatively, a combination of deterministic and stochastic mathematical methods were used to find the optimum size and binding affinity for nanoparticle penetration into a tumour76. Such approaches demonstrate the feasibility of integrating in silico methods with nanoparticle design. Other work has also explored broad biodistribution and tissue distribution of drug-antibody conjugates78,79. Here, simulations compared the difference in transport profiles between small molecules and larger macromolecular drugs. Four fundamental classes of drug delivery agents were highlighted (those limited by blood flow, vessel permeability, interstitial diffusion, and local binding and metabolism) each of which have strengths and weaknesses.
Along with optimising, in silico models have sought to explain why, in some instances, novel drug delivery methods lead to better accumulation and penetration than free-drug alternatives. For example, a general mathematical model was developed to investigate why experimental data showed a 3-fold drop in tumour growth when using nano-vectored drug delivery43. This mathematical model combined the cell cycle, vasculature network, and the drug diffusion rates, and was validated using in vivo methods. It demonstrated improved outcome for nanoparticle-based therapies, thought to be driven by the geometry of the particles. In vitro models have also confirmed improved tissue penetration and retention of nanoparticle-based therapies17,80. These improvements are thought to be in part due to tumour-scale phenomena such as the vascular network, which prevents certain cells from being reached. Hence, the combination of therapy with vasculature normalisation may provide a further improvement in treatment efficacy.
Endocytosis (END) and intracellular transport (IT)
Endocytosis can be performed by various cellular uptake mechanisms46,81,82. The rate and mechanism of cellular uptake depends on nanoparticle geometry, coating, and other physicochemical properties. Yet the specific design of a nanoparticle can greatly improve efficacy. Nanoparticles can be designed to increase cellular uptake (endocytosis) as well as to improve specificity by preferentially targeting cancer cells (through targeting receptors which are found to be upregulated in various tumour cells). Most in silico approaches consider a single nanoparticle or several (no more than ten) nanoparticles on the cell membrane and use either atomistic simulations or coarse-grain approaches such as DPD.
These studies have shown that the size and shape83, ligand multi-valency47 and the role of the protein corona around nanoparticles45 can all have considerable influence on cellular uptake. For example, super-selectivity was found to occur in multivalent nanoparticles, as they demonstrate ‘on-off’ binding profiles that is particularly well suited for receptor-concentration selective targeting47. This super-selectivity occurs when the fraction of bound particles sharply increases with receptor concentration. In this work, an analytical model was developed and compared against Monte Carlo simulations of nanoparticles with various coatings and general design principles were derived, once again demonstrating the use of in silico models in nanoparticle design. Other work has considered a thermodynamic model for nanoparticle–cell interactions84 where uptake rates were shown to be strongly related to the size of the nanoparticle and the interaction of multiple nanoparticles with the cell membrane85. Both works used coarse-grain approaches. For a comprehensive review of nanoparticle endocytosis see work by Angioletti-Uberti, and Zhang et al.46,49.
Having successfully been taken up by the cell, nanoparticles then face the challenge of intracellular transport (see Fig. 4). Post-endocytosis, nanoparticles can be localised at four major intracellular organelles: the cytoplasm, mitochondria, nucleus and lysosome39. Nanoparticles inside a cell face the risk of degradation by lysosomes. There are several means of overcoming this risk such as taking advantage of low intra-endosomal pH-induced osmotic swelling and endosome rupture (the proton sponge effect)38,86, or by hydrophilic to hydrophobic transition and endosomal membrane destabilisation induced by pH-sensitive amphiphilic polymeric nanoparticles17,38,87. Alternatively, a nanoparticle may be required to travel through the cytoplasm for highest effectiveness. However, the diffusion of nanoparticles within the cytoplasm can be significantly reduced due to binding with intracellular objects or the geometry of the nanoparticle. Currently, there are no in silico models that deal with intracellular transport at this level of detail.
Molecular dynamics (MD) simulations allow for in silico modelling of cellular uptake and intracellular trafficking of nanoparticles. Such models often give additional data where it is otherwise difficult to monitor the interaction of nanoparticles into and through a cell36. For example, MD simulations were used to show that hydrophobic nanoparticles had preferential entry to the endosomal pathway via the caveolae88. However, it is expected that additional data is required before models can be improved. Such data would allow for nanoparticles that are engineered with specific physicochemical properties that optimise intracellular transport and localisation in the areas where they finally induce their therapeutic effect. Studies considering nanoparticle geometry and surface functionality have been performed89. However, there is some way to go before more general causative effects can be pinpointed. This is further frustrated by the dynamic property of interactions between nanoparticles and intracellular objects and the fact that nanoparticles are often small enough to interact with one or many intracellular regions, simultaneously.
Currently available tools
We have discussed the biological barriers that prevent nanoparticle-based drug delivery from effectively treating tumours. In silico models allow for a more efficient means of exploring the nanoparticle parameter space than is possible with in vivo and in vitro models. However, the combination of nanoparticle behaviour within relevant biological models depends on the collaboration of multiple fields of research, including mathematical modellers, experimentalists, and clinicians. As such, free open-source software can streamline the design process by reducing model development time and allow more time for discussions on the most important model features that need to be included.
In Table 1, we give a summary of the tools, highlighting which transport barrier they can be used to address, model features such as the dimensionality, type of modelling framework, the scale and whether nanoparticles or proliferation have to be included, and if the codebase is open-source. We discuss some of these simulation tools in more detail below.
Ultimately, the choice of simulation software will depend on the tumour scenario that is being investigated, as several platforms are specifically designed to describe and address physiological scenarios. One example of this is the recent extension to the popular CHASTE system. CHASTE (cancer, heart, and soft-tissue environment) was designed with the specific goal of being a multi-purpose library for computational simulations of biological problems. The cancer codebase takes the cell-cycle of an individual cell as its lowest unit, allowing the user to alter parameters such as rates of cell division or the oxygen uptake. The cell-cycle model is connected to a tissue cell model which gives the user control of cell functionality including age, type, generation, mutational status and mitosis/apoptosis. The CHASTE library was introduced with functionality for producing both 2D crypts of tumour cells as well as 3D spheroid tumours90. More recently, the ability to model vascular transport has led to the publication of a library extension known as Microvessel CHASTE33.
Microvessel CHASTE allows for integration of 3D datasets of vascularised tissue to be incorporated into the simulations for comparison between in vitro/vivo and computational simulations. Microvessel CHASTE is able to model microvascular related phenomena including angiogenesis, tumour growth, and osteogenesis. Other computational fluid dynamics packages also capture behaviour within the vascular network but, as they are not designed specifically for biologically realistic environments, they fall out of the scope for this review. For further information on fluid dynamics as applied to oncology, see for example work by Koumoutsakos et al.66.
CHASTE and the subsequent Microvessel CHASTE are examples of combining clinical 3D datasets with computational simulations. There is an increasing number of libraries that offer extensive multi-scale functionality at the point of use, including CompuCell3D, Virtual Cell (or VCell), and PhysiCell. CompuCell3D is capable of modelling cell-scale to tissue-scale behaviour. It has been used to describe morphogenesis as well as cell-cell interactions, the immune response to cancer and spatial models of drug delivery routes for cancer treatment91,92. Similarly, VCell covers a wider range of modelling approaches which include continuum approaches (ODEs and PDEs), discrete models, and network models (for modelling intracellular signal pathways). It offers active integration with SpringSaLaD93, a simulation platform for modelling mesoscopic systems, but it is not currently open-source. PhysiCell94 is another multi-cellular library that implements hybrid methods to model 2D and 3D biological systems. It uses agent-based methods at the individual cell level and combines this with BioFVM95, a finite volume method specifically designed for biological systems. PhysiCell has been used to model tumours including growth and metastasis95,96 and extended to consider the signalling pathways through combination of the PhysiCell library with MaBoSS (used to run stochastic simulations of the signalling pathway), to create PhysiBoSS97.
Many questions on the effectiveness of nanoparticle-based therapies will depend on the model of interaction between nanoparticles and individual cells. To answer these questions, computational simulations are required that consider individual nanoparticle and their interactions, which ultimately change their dynamics. A computational framework has previously been suggested that specifically considers nanoparticle-scale interactions using both deterministic and stochastic methods76. These models used the stochastic simulation compiler (SSC) and led to the NanoDoc platform, a means of crowd-sourcing novel nanoparticle designs through online engagement (http://nanodoc.org). One of the most popular purpose-built libraries for modelling intracellular signal pathways is Smoldyn98, an agent-based model that can effectively describe nanosecond to microsecond molecular-resolution behaviour. This package is open-source and has been used to describe macromolecular crowding and molecular diffusion. Similarly, the stochastic engine for pathway simulation (STEPS) is an open source package for stochastic simulations of reaction-diffusion systems where the geometry of the domain is important99. Originally built to simulate neuronal signalling pathways, the length-scales consider by this package make it a viable tool for nanoparticle research. Furthermore, STEPS has been shown to effectively compute large-scale simulations using multi-core parallelisation, increasing the size of simulated systems100. To the best of the authors' knowledge, neither Smolydn nor STEPS have yet to be applied specifically to nanoparticle simulation. Finally, there are several molecular dynamic simulation tools that are available which have been used to consider nanoparticle design. While not specific to nanomedicine or oncology, these have been reviewed in, for example, work by Cummings et al.101.
Machine learning and data-driven approaches
All the above discussions have centred on the overlap between in silico models of nanoparticle transport barriers. The current state of the field within nanomedicine is similar to that seen in material sciences, biology, and medicine where multi-scale simulations have led to a far wider exploration of system dynamics than possible prior to increases in computational power and data. However, in recent years the application of machine learning and data-driven methods has led to a significant shift in research methodology and contributed to advances in these (as well as many other) fields18,102,103. Moreover, it is expected that the application of these methodologies will accelerate in the future. Hence, it is important to characterise these techniques in relation to nanomedicine.
Describing the current state-of-the-art in machine learning is beyond the scope of this review and has been well described in other work104,105. Furthermore, while the application of machine learning tools to the field of nanomedicine is still relatively uncommon, these methods have already been applied specifically to characterisation of drug-loaded NPs. For example, artificial neural networks have been used to predict drug-loaded NP size and polydispersity106,107, both having clear implications for the efficacy of nanoparticle-based treatments. Other work has used machine learning tools to optimise cellular uptake, minimise cytotoxicity and predict the development of the protein corona around a nanoparticle108,109,110,111. For a recent review of this area, see for example, work by Jones et al. and Sason et al.112,113. Ultimately, these techniques further benefit from their integration with in silico models, as this leads to a speed up in time and reduction in costs, as output from in silico models become input for machine learning techniques. This has already been seen in material sciences, where algorithmic learning of specific models can massively speed up the exploration of a system’s state space102.
Whereas the characterisation and design of nanoparticles is expected to greatly benefit from utilisation of machine learning methods, additional benefits can also be gained from the combination of machine learning with experiment design. Active learning uses machine learning techniques (such as reinforcement learning114 or surrogate-assisted optimisation115) to guide experiment design by selecting for only the most promising candidates that require testing. This approach, distinct from traditional trial-and-error, has already shown promise within the fields of clinical cancer trials, drug discovery and material discovery18,116,117. We believe that designing nanoparticles to overcome transport barriers is a rich future area of study118. Furthermore, the integration of in silico models with active learning will allow for both the automatic exploration and nanoparticle designs, and a way to test them, often a pre-requisite when implementing an active learning approach102.
With recent advances in multi-scale simulations of tumours and nanomedicines and their combination with machine learning techniques, the application of in silico methods in a clinical setting is beginning to become reality. However, establishing the clinical relevance of computational models requires industry-grade evidence gained through verification and validation, sensitivity analysis, and uncertainty quantification, amongst other things119. The level of rigour of these evidences will depend on both their intended use and the clarity of such use at the outset. Hence, the existence of guidance documents is critical in the successful and safe application of such methods to the clinical setting120. In 2013, the International Medical Device Regulators Forum (IMDRF) published the first in a series of guidance documents on ‘Software as a Medical Device’ (SaMD). They covered key definitions121, categorisation122, quality management system123 and planning the process for clinical evaluation of a SaMD124.
Taken together those documents provide a risk-based framework where the level of clinical evidence depends on the risk profile of a SaMD, where the risk profile is characterised by the severity of the underlying health condition and the significance of the information provided by the SaMD to the healthcare decision maker. Since computational models of physiological systems are usually nonlinear, highly complex, and contain high numbers of parameters and time-dependent properties, their proper assessment can be very challenging. On top of this, the introduction of machine learning into simulation models opens up a new set of problems including lack of transparency, automation bias (where there is a tendency of users to non-critically accept computer recommendations) and changes in input/output relations as an algorithm learns from real-world use and experience. In 2019, the FDA published a discussion paper which calls for medical machine learning algorithms to include the types of anticipated modifications125. In light of this, we advise developers, as early as possible in the development pipeline, to get familiar with the possible risks associated with their simulation platform and take into account relevant sources of evidence (such as verification, validation, and uncertainty quantification) that are likely to be required in clinical settings.
Conclusions and future perspective
Overcoming nanoparticle transport barriers, specifically travel through the vasculature, extravasation, tissue penetration, endocytosis and the delivery of a therapeutic cargo, is paramount to the effective use of nanoparticles or anti-cancer treatments. In silico tools allow for the fast and systematic exploration of the nanoparticle design space to select nanoparticles with the potential to deliver their cargo to the right place. General guidelines extracted from such tools could prove useful in making more effective treatments. Specific solutions could also be provided to tailor nanoparticles to patient needs and towards personalised medicine, or to produce sufficient amounts of data for machine learning. Building useful in silico tools will require close validation with in vitro and in vivo results. The overarching aim is to create a systems approach to nanomedicine which accounts for multi-scale phenomena, which is repeatedly validated, and which exists within a shared modelling framework. We describe these elements in more detail below.
To become effective, nanoparticle cancer therapies require a systematic approach to prototyping. In silico modelling has now advanced to the point of being an effective tool that can minimise costly trial-and-error design methods (see, for example, the review by Karolak et al.50). However, such modelling cannot exist in isolation and a collaboration between mathematical modellers, experimentalists, and clinicians will be required to inform the best transfer of knowledge. As a first step, this method will help identify guidelines for the design of suitable nanoparticles for a class of problems, say a specific tumour type. In the future, integration of patient data such as MRI scans or biopsies can inform computational models of tumour growth and offer a route to personalised nanoparticles126,127. We envisage a pipeline where theoretical predictions are checked against clinical outcomes and then returned to inform future simulations, as shown in Fig. 5.
Tumour progression and nanoparticle transport exhibit clear multi-scale behaviour. This can be understood within a systems biology framework128,129; individual microscopic effects are more than the sum of their parts when viewed in aggregate. It is not straightforward to determine the probability distribution of emergent behaviour using microstate characterisation, as the variables in complex biological systems are interdependent and unlikely to be normally distributed27. Yet, by testing multiple in silico models, a sufficient sampling of the model outputs allows us to project microstate knowledge higher up the hierarchy of scales. This is possible using in silico models as system parameters can be closely controlled for and many thousands of possible model scenarios can be investigated in a systematic and efficient way. Using a systems approach to cancer modelling, covering topics such as tissue complexity, cell heterogeneity, targeted therapy, and drug resistance, has been reviewed in, for example, work by Werner et al.130.
This system approach highlights three immediate advantages of using multi-scale in silico models: improved hypothesis testing, strategy generation, and clinical relevance. Brute force simulation experiments allow for much wider characterisation and sampling of a systems state-space73. These allows the simulations to test whether microstate phenomena have a verifiable causal relation to emergent macroscale observables. This is the hypothesis testing benefit. For example, PhysiCell has been used to run large-scale parallelised simulations that generate data-driven error metrics and create in silico validations of biomedical hypotheses131.
Second, the increased sample rate gained from running many thousands of simultaneous simulations generates much larger data. Increased data, combined with optimisation and machine learning, will lead to novel, more efficient, strategy generation76. Furthermore, by combining computer simulations with patient data and experimental results, these novel strategies can be implemented and checked to explore new medical applications.
This motivates the third advantage of silico models. In silico models validated and optimised by in vivo and/or in vitro models have both better explanatory and predictive power. The strength of multi-scale in silico models is the ability to bridge length-scales which have traditionally isolated causal factors. By integrating across scales, the predictive power of in silico modelling approaches can be increased with a corresponding expected payoff to clinical relevance. Furthermore, the combination of simulating across scales with parameters gained from in vitro and/or in vivo can expand causal relations beyond scale of observations (such as relating intercellular growth models to the observed change in the size of a tumour). Finally, the combination of these methods with machine learning techniques can be used to build causal models from correlations inferred from big data103.
The three advantages of in silico models (improved hypothesis testing, strategy generation, and clinical relevance) will require standardisation between computational models such that relevant benchmarks can be used to compare various in silico approaches. Furthermore, a common design framework which includes shared datatypes will allow for expedited comparison between models that have a shared common input. This has been advocated before132,133,134, but only time will tell whether such a standardisation becomes successful.
Data availability
All relevant data are available from the authors.
References
Etheridge, M. L. et al. The big picture on nanomedicine: the state of investigational and approved nanomedicine products Nanomedicine 9, 1–14 (2013).
Su, Y. L. & Hu, S. H. Functional nanoparticles for tumor penetration of therapeutics. Pharmaceutics 10, 1–21 (2018).
Tran, S., DeGiovanni, P.-J., Piel, B. & Rai, P. Cancer nanomedicine: a review of recent success in drug delivery. Clin. Transl. Med. 6, 44 (2017).
Hare, J. I. et al. Challenges and strategies in anti-cancer nanomedicine development: an industry perspective. Adv. Drug Deliv. Rev. 108, 25–38 (2017).
Strambeanu, N., Demetrovici, L., Dragos, D. & Lungu, M. in Nanoparticles’ Promises and Risks: Characterization, Manipulation, and Potential Hazards to Humanity and the Environment (eds Lungu, M., Neculae, A., Bunoiu, M. & Biris, C.) (Springer, 2015).
Roberts, W. G. & Palade, G. E. G. Increased microvascular permeability and endothelial fenestration induced by vascular endothelial growth factor. J. Cell Sci. 108, 2369–2379 (1995).
Tong, R., Hemmati, H. D., Langer, R. & Kohane, D. S. Photoswitchable nanoparticles for triggered tissue penetration and drug delivery. J. Am. Chem. Soc. 134, 8848–8855 (2012).
Kong, S. D. et al. Magnetically vectored nanocapsules for tumor penetration and remotely switchable on-demand drug release. Nano Lett. 10, 5088–5092 (2010).
Wang, B. et al. Simultaneously overcome tumor vascular endothelium and extracellular matrix barriers via a non-destructive size-controlled nanomedicine. J. Control. Release 268, 225–236 (2017).
Bazak, R. et al. Cancer active targeting by nanoparticles: a comprehensive review of literature. J. Cancer Res. Clin. Oncol. 141, 769–784 (2015).
Hauert, S. & Bhatia, S. N. Mechanisms of cooperation in cancer nanomedicine: towards systems nanotechnology. Trends Biotechnol. 32, 448–455 (2014).
Li, Y. et al. Cell and nanoparticle transport in tumour microvasculature: the role of size, shape and surface functionality of nanoparticles. Interface Focus 6, 1–15 (2016).
Park, J.-H. et al. Cooperative nanomaterial system to sensitize, target, and treat tumors. Proc. Natl Acad. Sci. 107, 981–986 (2010).
Von Maltzahn, G. et al. Nanoparticles that communicate in vivo to amplify tumour targeting. Nat. Mater. 10, 545–552 (2011).
Fu, Y. et al. A feasible strategy for self-assembly of gold nanoparticles: via dithiol-PEG for photothermal therapy of cancers. RSC Adv. 8, 6120–6124 (2018).
Xiao, Z. et al. DNA self-assembly of targeted near-infrared-responsive gold nanoparticles for cancer thermo-chemotherapy. Angew. Chem.—Int. Edn. 51, 11853–11857 (2012).
Bae, Y. H. & Park, K. Targeted drug delivery to tumors: myths, reality and possibility. J. Control. Release 153, 198–205 (2011).
Lookman, T., Balachandran, P. V., Xue, D. & Yuan, R. Active learning in materials science with emphasis on adaptive sampling using uncertainties for targeted design. npj Comput. Mater. 5, 1–17 (2019).
Deisboeck, T. S., Zhang, L., Yoon, J. & Costa, J. In silico cancer modeling: is it ready for prime time? Nat. Clin. Practice Oncol. 6, 34–42 (2009).
Michor, F. & Beal, K. Improving cancer treatment via mathematical modeling: surmounting the challenges is worth the effort. Cell 163, 1059–1063 (2015).
Dogra, P. et al. Mathematical modeling in cancer nanomedicine: a review. Biomed. Microdevices 21, 40 (2019).
Talmadge, J. E. & Fidler, I. J. The biology of cancer metastasis: historical perspective. Cancer Res. 70, 5649–5669 (2010).
Siegel, R., Miller, K. D. & Ahmedin, J. Cancer Statistics, 2017. CA: Cancer J. Clinicians 67, 7–30 (2017).
Chauviere, A. H. et al. Mathematical oncology: how are the mathematical and physical sciences contributing to the war on breast cancer? Curr. Breast Cancer Rep. 2, 121–129 (2010).
Deisboeck, Z. & Yoon, C. In silico modelling—is it ready for prime time. Program 6, 34–42 (2011).
Rejniak, K. A. & Anderson, A. R. Hybrid models of tumor growth. Wiley Interdisciplinary Rev.: Syst. Biol. Med. 3, 115–125 (2011).
An, G. & Mi, Q. Agent based models in translational systems biology. Syst. Biol. Med. 1, 159–171 (2009).
Deisboeck, T. S. & Stamatakos, G. S. Multiscale Cancer Modeling (CRC Press, 2010).
Norton, K.-A., Gong, C., Jamalian, S. & Popel, A. Multiscale agent-based and hybrid modeling of the tumor immune microenvironment. Processes 7, 1–23 (2019).
Metzcar, J., Wang, Y., Heiland, R. & Macklin, P. A review of cell-based computational modeling in cancer biology. JCO Clin. Cancer Inform. 3, 1–13 (2019).
Zhu, X., Zhou, X., Lewis, M. T., Xia, L. & Wong, S. Cancer stem cell, niche and EGFR decide tumor development and treatment response: a bio-computational simulation study. J. Theor. Biol. 269, 138–49 (2011).
Sefidgar, M. et al. Numerical modeling of drug delivery in a dynamic solid tumor microvasculature. Microvascular Res. 99, 43–56 (2015).
Grogan, J. A. et al. Microvessel chaste: an open library for spatial modeling of vascularized tissues. Biophys. J. 112, 1767–1772 (2017).
Shah, P. N. et al. Extravasation of Brownian spheroidal nanoparticles through vascular pores. Biophys. J. 115, 1103–1115 (2018).
Owen, M. R. et al. Mathematical modeling predicts synergistic antitumor effects of combining a macrophage-based, hypoxia-targeted gene therapy with chemotherapy. Cancer Res. 71, 2826–2837 (2011).
Chou, C. Y., Huang, C. K., Lu, K. W., Horng, T. L. & Lin, W. L. Investigation of the spatiotemporal responses of nanoparticles in tumor tissues with a small-scale mathematical model. PLoS ONE 8, e59135 (2013).
Nehoff, H., Parayath, N. N., Domanovitch, L., Taurin, S. & Greish, K. Nanomedicine for drug targeting: strategies beyond the enhanced permeability and retention effect. Int. J. Nanomed. 9, 2539–55 (2014).
Chrastina, A., Massey, K. A. & Schnitzer, J. E. Overcoming in vivo barriers to targeted nanodelivery. Wiley Interdisciplinary Rev.: Nanomed. Nanobiotechnol. 3, 421–37 (2011).
Barua, S. & Mitragotri, S. Challenges associated with penetration of nanoparticles across cell and tissue barriers: a review of current status and future prospects. Nano Today 9, 223–243 (2014).
Curtis, L. T., England, C. G., Wu, M., Lowengrub, J. & Frieboes, H. B. An interdisciplinary computational/experimental approach to evaluate drug-loaded gold nanoparticle tumor cytotoxicity. Nanomedicine 11, 197–216 (2016).
Shah, A. B., Rejniak, K. A. & Gevertz, J. L. Limiting the development of anti-cancer drug resistance in a spatial model of micrometastases. Math. Biosci. Eng. 13, 1185–1206 (2016).
Al-Obaidi, H. & Florence, A. T. Nanoparticle delivery and particle diffusion in confined and complex environments. J. Drug Deliv. Sci. Technol. 30, 266–277 (2015).
Wang, Z. et al. Theory and experimental validation of a spatio-temporal model of chemotherapy transport to enhance tumor cell kill. PLoS Comput. Biol.12, e1004969 (2016).
Hamis, S., Nithiarasu, P. & Powathil, G. G. What does not kill a tumour may make it stronger: In silico insights into chemotherapeutic drug resistance. J. Theor. Biol. 454, 253–267 (2018).
ming Ding, H. & qiang Ma, Y. Computer simulation of the role of protein corona in cellular delivery of nanoparticles. Biomaterials 35, 8703–8710 (2014).
Zhang, S., Gao, H. & Bao, G. Physical principles of nanoparticle cellular endocytosis. ACS Nano 9, 8655–8671 (2015).
Martinez-Veracoechea, F. J. & Frenkel, D. Designing super selectivity in multivalent nano-particle binding. Proc. Natl Acad. Sci. 108, 10963–10968 (2011).
Pascal, J. et al. Mechanistic modeling identifies drug-uptake history as predictor of tumor drug resistance and nano-carrier-mediated response. ACS Nano 7, 11174–11182 (2013).
Angioletti-Uberti, S. Theory, simulations and the design of functionalized nanoparticles for biomedical applications: a soft matter perspective. npj Comput. Mater. 3, 1–48 (2017).
Karolak, A., Markov, D. A., McCawley, L. J. & Rejniak, K. A. Towards personalized computational oncology: from spatial models of tumour spheroids, to organoids, to tissues. J. Roy. Soc. Interface 15, 20170703 (2018).
Afonin, K. A. et al. In silico design and enzymatic synthesis of functional RNA nanoparticles. Acc. Chem. Res. 47, 1731–1741 (2014).
Finley, S. D., Angelikopoulos, P., Koumoutsakos, P. & Popel, A. S. Pharmacokinetics of Anti-VEGF Agent aflibercept in cancer predicted by data-driven, molecular-detailed model. CPT: Pharmacometrics Syst. Pharmacol. 4, 641–649 (2015).
Wang, Z., Bordas, V., Sagotsky, J. & Deisboeck, T. S. Identifying therapeutic targets in a combined EGFR-TGFβ R signalling cascade using a multiscale agent-based cancer model. Math. Med. Biol. 29, 95–108 (2012).
Blanco, E., Shen, H. & Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nature Biotechnol. 33, 9 (2015).
Longmire, M., Choyke, P. L. & Kobayashi, H. Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine 3, 703–717 (2008).
Gustafson, H. H., Holt-Casper, D., Grainger, D. W. & Ghandehari, H. Nanoparticle uptake: the phagocyte problem. Nano Today 10, 487–510 (2015).
Fedosov, D. A., Noguchi, H. & Gompper, G. Multiscale modeling of blood flow: from single cells to blood rheology. Biomech. Model. Mechanobiol. 13, 239–258 (2014).
Lopez, H. & Lobaskin, V. Coarse-grained model of adsorption of blood plasma proteins onto nanoparticles. J. Chem. Phys. 143, 12B620_1 (2015).
Shao, Q. & Hall, C. K. Protein adsorption on nanoparticles: model development using computer simulation. J. Phys.: Condens. Matter 28, 414019 (2016).
Maleki, R. et al. ph-sensitive loading/releasing of doxorubicin using single-walled carbon nanotube and multi-walled carbon nanotube: a molecular dynamics study. Comput. Methods Programs Biomed. 186, 105210 (2020).
Yoo, J.-W., Chambers, E. & Mitragotri, S. Factors that control the circulation time of nanoparticles in blood: challenges, solutions and future prospects. Curr. Pharm. Des. 16, 2298–2307 (2010).
Müller, K., Fedosov, D. A. & Gompper, G. Margination of micro- and nano-particles in blood flow and its effect on drug delivery. Sci. Rep. 4, 1–8 (2014).
Lin, Z., Monteiro-Riviere, N. A. & Riviere, J. E. A physiologically based pharmacokinetic model for polyethylene glycol-coated gold nanoparticles of different sizes in adult mice. Nanotoxicology 10, 162–172 (2016).
Yuan, D., He, H., Wu, Y., Fan, J. & Cao, Y. Physiologically based pharmacokinetic modeling of nanoparticles. J. Pharm. Sci. 108, 58–72 (2019).
Ding, H.-m & Ma, Y.-q Computational approaches to cell–nanomaterial interactions: keeping balance between therapeutic efficiency and cytotoxicity. Nanoscale Horizons 3, 6–27 (2017).
Koumoutsakos, P., Pivkin, I. & Milde, F. The fluid mechanics of cancer and its therapy. Annu. Rev. Fluid Mech. 45, 325–355 (2013).
Nakamura, Y., Mochida, A., Choyke, P. L. & Kobayashi, H. Nanodrug delivery: is the enhanced permeability and retention effect sufficient for curing cancer? Bioconjugate Chem. 27, 2225–2238 (2016).
Björnmalm, M., Thurecht, K. J., Michael, M., Scott, A. M. & Caruso, F. Bridging bio-nano science and cancer nanomedicine. ACS Nano 11, 9594–9613 (2017).
Nichols, J. W. & Bae, Y. H. Odyssey of a cancer nanoparticle: from injection site to site of action. Nano Today 7, 606–618 (2012).
Fullstone, G., Wood, J., Holcombe, M. & Battaglia, G. Modelling the transport of nanoparticles under blood flow using an agent-based approach. Sci. Rep. 5, 1–13 (2015).
Ferretti, S., Allegrini, P. R., Becquet, M. M. & McSheehy, P. M. Tumor interstitial fluid pressure as an early-response marker for anticancer therapeutics. Neoplasia 11, 874–881 (2015).
Lameijer, M. A., Tang, J., Nahrendorf, M., Beelen, R. H. & Mulder, W. J. Monocytes and macrophages as nanomedicinal targets for improved diagnosis and treatment of disease. Expert Rev. Mol. Diagnostics 13, 576–580 (2013).
Wu, M. et al. The effect of interstitial pressure on therapeutic agent transport: coupling with the tumor blood and lymphatic vascular systems. J.Theor. Biol. 335, 194–207 (2014).
Wijeratne, P. A. & Vavourakis, V. A quantitative in silico platform for simulating cytotoxic and nanoparticle drug delivery to solid tumours. Interface Focus 9, 20180063 (2019).
Frieboes, H. B., Wu, M., Lowengrub, J., Decuzzi, P. & Cristini, V. A computational model for predicting nanoparticle accumulation in tumor vasculature. PLoS ONE 8, 1–11 (2013).
Hauert, S., Berman, S., Nagpal, R. & Bhatia, S. N. A computational framework for identifying design guidelines to increase the penetration of targeted nanoparticles into tumors. Nano Today 8, 566–576 (2013).
Chamseddine, I. M., Frieboes, H. B. & Kokkolaras, M. Design optimization of tumor vasculature-bound nanoparticles. Sci. Rep. 8, 17768 (2018).
Thurber, G. M. & Weissleder, R. A systems approach for tumor pharmacokinetics. PLoS ONE 6, e24696 (2011).
Cilliers, C., Guo, H., Liao, J., Christodolu, N. & Thurber, G. M. Multiscale modeling of antibody-drug conjugates: Connecting tissue and cellular distribution to whole animal pharmacokinetics and potential implications for efficacy. The AAPS J. 18, 1117–1130 (2016).
Liu, J. et al. Design of nanocarriers based on complex biological barriers in vivo for tumor therapy. Nano Today 15, 56–90 (2017).
Daum, N., Tscheka, C., Neumeyer, A. & Schneider, M. Novel approaches for drug delivery systems in nanomedicine: Effects of particle design and shape. Wiley Interdisciplinary Rev.: Nanomed. Nanobiotechnol. 4, 52–65 (2012).
Anselmo, A. C. et al. Elasticity of nanoparticles influences their blood circulation, phagocytosis, endocytosis, and targeting. ACS Nano 9, 3169–3177 (2015).
Bao, G. et al. USNCTAM perspectives on mechanics in medicine. J. Roy. Soc. Interface 11, 20140301 (2014).
Zhang, S., Li, J., Lykotrafitis, G., Bao, G. & Suresh, S. Size-dependent endocytosis of nanoparticles. Adv. Mater. 21, 419–424 (2009).
Yue, T. & Zhang, X. Cooperative effect in receptor-mediated endocytosis of multiple nanoparticles. ACS Nano 6, 3196–3205 (2012).
Stewart, M. P., Lorenz, A., Dahlman, J. & Sahay, G. Challenges in carrier-mediated intracellular delivery: moving beyond endosomal barriers. Wiley Interdisciplinary Rev.: Nanomed. Nanobiotechnol. 8, 465–478 (2016).
Martens, T. F., Remaut, K., Demeester, J., De Smedt, S. C. & Braeckmans, K. Intracellular delivery of nanomaterials: How to catch endosomal escape in the act. Nano Today 9, 344–364 (2014).
Chiu, Y.-L. et al. The characteristics, cellular uptake and intracellular trafficking of nanoparticles made of hydrophobically-modified chitosan. J. Control. Release 146, 152–159 (2010).
Chithrani, B. D. & Chan, W. C. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 7, 1542–1550 (2007).
Pitt-Francis, J. et al. Chaste: a test-driven approach to software development for biological modelling. Comput. Phys. Commun. 180, 2452–2471 (2009).
Winner, K. R. et al. Spatial modeling of drug delivery routes for treatment of disseminated ovarian cancer. Cancer Res. 76, 1320–1334 (2016).
Li, J. F. & Lowengrub, J. The effects of cell compressibility, motility and contact inhibition on the growth of tumor cell clusters using the Cellular Potts Model. J. Theor. Biol. 343, 79–91 (2014).
Michalski, P. J. & Loew, L. M. SpringSaLaD: a spatial, particle-based biochemical simulation platform with excluded volume. Biophys. J. 110, 523–529 (2016).
Ghaffarizadeh, A., Heiland, R., Friedman, S. H., Mumenthaler, S. M. & Macklin, P. PhysiCell: an open source physics-based cell simulator for 3-D multicellular systems. PLoS Comput. Biol. 14, e1005991 (2018).
Ghaffarizadeh, A., Friedman, S. H. & MacKlin, P. BioFVM: an efficient, parallelized diffusive transport solver for 3-D biological simulations. Bioinformatics 32, 1256–1258 (2016).
Juarez, E. F., Garri, C., Ghaffarizadeh, A., Macklin, P. & Kani, K. Quantification of cancer cell migration with an integrated experimental-computational pipeline. F1000Research 7, 1296 (2018).
Letort, G. et al. PhysiBoSS: a multi-scale agent-based modelling framework integrating physical dimension and cell signalling. Bioinformatics 35, 1188–1196 (2019).
Andrews, S. S., Addy, N. J., Brent, R. & Arkin, A. P. Detailed simulations of cell biology with Smoldyn 2.1. PLoS Comput. Biol. 6, e1000705 (2010).
Hepburn, I., Chen, W., Wils, S. & De Schutter, E. STEPS: efficient simulation of stochastic reaction-diffusion models in realistic morphologies. BMC Syst. Biol. 6, 36 (2012).
Chen, W. & De Schutter, E. Parallel STEPS: large scale stochastic spatial reaction-diffusion simulation with high performance computers. Frontiers Neuroinform. 11, 1–15 (2017).
Cummings, P. T. & Gilmer, J. B. Open-source molecular modeling software in chemical engineering. Curr. Opin. Chem. Eng. 23, 99–105 (2019).
Schmidt, J., Marques, M. R., Botti, S. & Marques, M. A. Recent advances and applications of machine learning in solid-state materials science. npj Comput. Mater. 5, 1–36 (2019).
Alber, M. et al. Integrating machine learning and multiscale modeling-perspectives, challenges, and opportunities in the biological, biomedical, and behavioral sciences. npj Digital Med. 2, 1–11 (2019).
Bishop, C. M. Pattern Recognition and Machine Learning (springer, 2006).
Alpaydin, E. Introduction to Machine Learning (MIT press, 2020).
Asadi, H., Rostamizadeh, K., Salari, D. & Hamidi, M. Preparation of biodegradable nanoparticles of tri-block pla–peg–pla copolymer and determination of factors controlling the particle size using artificial neural network. J. Microencapsulation 28, 406–416 (2011).
Shalaby, K. S. et al. Determination of factors controlling the particle size and entrapment efficiency of noscapine in peg/pla nanoparticles using artificial neural networks. Int. J. Nanomed. 9, 4953 (2014).
Liu, R. et al. Classification nanosar development for cytotoxicity of metal oxide nanoparticles. Small 7, 1118–1126 (2011).
Hataminia, F., Noroozi, Z. & Eslam, H. M. Investigation of iron oxide nanoparticle cytotoxicity in relation to kidney cells: a mathematical modeling of data mining. Toxicol. in Vitro 59, 197–203 (2019).
Labouta, H. I., Asgarian, N., Rinker, K. & Cramb, D. T. Meta-analysis of nanoparticle cytotoxicity via data-mining the literature. ACS Nano 13, 1583–1594 (2019).
Findlay, M. R., Freitas, D. N., Mobed-Miremadi, M. & Wheeler, K. E. Machine learning provides predictive analysis into silver nanoparticle protein corona formation from physicochemical properties. Environ. Sci.: Nano 5, 64–71 (2018).
Jones, D. E., Ghandehari, H. & Facelli, J. C. A review of the applications of data mining and machine learning for the prediction of biomedical properties of nanoparticles. Comput. Methods Programs Biomed. 132, 93–103 (2016).
Sason, H. & Shamay, Y. Nanoinformatics in drug delivery. Israel J. Chem. https://doi.org/10.1002/ijch.201900042 (2019).
Sutton, R. S. & Barto, A. G. Reinforcement Learning: An Introduction (MIT press, 2018).
Jin, Y. Surrogate-assisted evolutionary computation: recent advances and future challenges. Swarm Evolut. Comput. 1, 61–70 (2011).
Zhao, Y., Kosorok, M. R. & Zeng, D. Reinforcement learning design for cancer clinical trials. Statistics Med. 28, 3294–3315 (2009).
Warmuth, M. K. et al. Active learning with support vector machines in the drug discovery process. J. Chem. Inform. Comput. Sci. 43, 667–673 (2003).
Yamankurt, G. et al. Exploration of the nanomedicine-design space with high-throughput screening and machine learning. Nature Biomed. Eng. 3, 318–327 (2019).
Parvinian, B. et al. Credibility evidence for computational patient models used in the development of physiological closed-loop controlled devices for critical care medicine. Frontiers Physiol. 10, 220 (2019).
Morrison, T. M. et al. Assessing computational model credibility using a risk-based framework: application to hemolysis in centrifugal blood pumps. ASAIO J. 65, 349 (2019).
Software as a Medical Device (SaMD): Key Definitions (2013). Available at: http://www.imdrf.org/docs/imdrf/final/technical/imdrf-tech-131209-samd-key-definitions-140901.pdf.
Software as a Medical Device- (SaMD): Possible Framework for Risk Categorization and Corresponding Considerations (2014). Available at: http://www.imdrf.org/docs/imdrf/final/technical/imdrf-tech-140918-samd-framework-risk-categorization-141013.pdf.
Software as a Medical Device- (SaMD): Application of Quality Management Systems. (2014). Available at: http://www.imdrf.org/docs/imdrf/final/technical/imdrf-tech-151002-samd-qms.pdf.
Software as a Medical Device- (SaMD): Clinical Evaluation (2017). Available at: http://www.imdrf.org/docs/imdrf/final/technical/imdrf-tech-170921-samd-n41-clinical-evaluation_1.pdf.
Proposed regulatory framework for modifications to artificial intelligence/machine learning (AI/ML)-based software as a medical device (SaMD) (2019). Available at: https://www.fda.gov/media/122535/download.
Weis, J. A. et al. Predicting the response of breast cancer to neoadjuvant therapy using a mechanically coupled reaction-diffusion model. Cancer Res. 10, 4333–4347 (2015).
Macklin, P., Edgerton, M. E., Thompson, A. M. & Cristini, V. Patient-calibrated agent-based modelling of ductal carcinoma in situ (DCIS): From microscopic measurements to macroscopic predictions of clinical progression. J. Theor. Biol. 301, 122–140 (2012).
Korsunsky, I. et al. Systems biology of cancer: a challenging expedition for clinical and quantitative biologists. Frontiers Bioeng. Biotechnol. 2, 27 (2014).
Faratian, D., Bown, J. L., Smith, V. A., Langdon, S. P. & Harrison, D. J. Cancer Systems Biology 245–263 (Humana Press, Totowa, NJ, 2010).
Werner, H. M., Mills, G. B. & Ram, P. T. Cancer systems biology: a peek into the future of patient care? Nat. Rev. Clin. Oncol. 11, 167–176 (2014).
Kim, M. Y. et al. Tumor self-seeding by circulating cancer cells. Cell 139, 1315–1326 (2009).
Koutsoukas, A. et al. From in silico target prediction to multi-target drug design: current databases, methods and applications. J. Proteomics 74, 254–2574 (2011).
Macklin, P. et al. Progress Towards Computational 3-D Multicellular Systems Biology 225–246 (Springer International Publishing, 2016).
Rockne, R. C. et al. The 2019 mathematical oncology roadmap. Physical Biology 16, 041005 (2019).
Acknowledgements
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 800983.
Author information
Authors and Affiliations
Contributions
All authors contributed to the content of the paper. N.S. led the creation and writing of the manuscript with input from M.K., I.B., and S.H. S.H. defined the structure and scope of the paper, supervised its writing, and provided feedback on all stages of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stillman, N.R., Kovacevic, M., Balaz, I. et al. In silico modelling of cancer nanomedicine, across scales and transport barriers. npj Comput Mater 6, 92 (2020). https://doi.org/10.1038/s41524-020-00366-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-020-00366-8
This article is cited by
-
Mathematical Modeling of Micro-/Nanoparticles Transport in Blood Vessels: A Review
Korean Journal of Chemical Engineering (2024)
-
An in silico model of the capturing of magnetic nanoparticles in tumour spheroids in the presence of flow
Biomedical Microdevices (2024)
-
Targeting cancer with mRNA–lipid nanoparticles: key considerations and future prospects
Nature Reviews Clinical Oncology (2023)
-
Predicting efficacy of drug-carrier nanoparticle designs for cancer treatment: a machine learning-based solution
Scientific Reports (2023)
-
Nanoparticle-mediated cancer cell therapy: basic science to clinical applications
Cancer and Metastasis Reviews (2023)