Only 12 years ago, an H1N1 influenza, which was detected first in the United States, spread quickly across the United States and the world. In one year, there were 60.8 million cases, 274,304 hospitalizations, and 12,469 deaths in the United States, as estimated by the CDC. Additionally, up to a half of a million people died worldwide during the first year the virus circulated.
A new study shows that during that pandemic, people developed strong, effective immune responses to stable, conserved parts of the virus. Protection against different strains of influenza, the authors wrote, is mediated by broadly neutralizing antibodies. The antibodies responding to the 2009 pandemic H1N1 influenza virus, they find, were broadly neutralizing and were specific to one of two conserved regions of the hemagglutinin (HA) head, the lateral patch or the receptor binding site (RBS). In addition, mice receiving passive transfer of these antibodies were protected from lethal influenza infection.
These findings suggest a strategy for developing universal flu vaccines that are designed to generate those same responses, instead of targeting parts of the virus that tend to evolve rapidly and require a new vaccine every year.
This work is published in Science Translational Medicine in the paper, “First exposure to the pandemic H1N1 virus induced broadly neutralizing antibodies targeting hemagglutinin head epitopes.”
Influenza is an elusive and frustrating target for vaccines. There are two main types of flu virus that can infect humans, which evolve rapidly from season to season. When developing seasonal flu vaccines, health officials try to anticipate the predominant variation of the virus that will circulate that year. Sometimes new, unexpected variants emerge, which means the vaccine may not be very effective. To avoid this, the ultimate goal of many flu researchers is to develop a universal vaccine that can account for any virus strain or variation in a given year, or even longer.
The new study, led by immunologists at the University of Chicago and structural biologists from Scripps Research Institute, studied the immune responses of people who were first exposed to the 2009 H1N1 pandemic flu virus, either from infection or a vaccine.
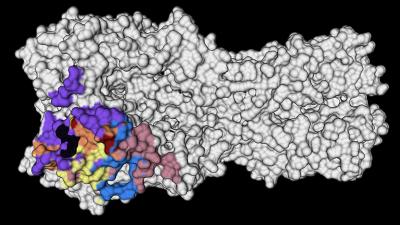
The researchers saw that the immune systems of these people recalled memory B cells from their childhood that produced broadly neutralizing antibodies against highly conserved parts on the head of the HA protein—a virus surface protein that attaches to receptors on host cells. Specifically, monoclonal antibodies (mAbs) generated against the conserved RBS or lateral patch epitopes of the HA head domain epitopes are broadly neutralizing against H1N1 viruses. The authors wrote that, “lateral patch-targeting antibodies demonstrated near universal binding to H1 viruses, and RBS-binding antibodies commonly cross-reacted with H3N2 viruses and influenza B viruses.”
Moreover, the data indicate that RBS- and lateral patch-targeting clones are abundant within the human memory B cell pool, and universal vaccine strategies should aim to drive antibodies against both conserved head and stalk epitopes.
In a separate 2020 study, Jenna Guthmiller, PhD, a post-doc in the lab of Patrick Wilson, PhD, professor of immunology at the University of Chicago, and colleagues, found polyreactive antibodies that can bind to several conserved sites on the flu virus. Now, the new study reveals more details about the conditions that can recall the same strong immune responses as this first exposure.
“That’s the exciting thing about this study,” Guthmilller said. “Not only have we found these broadly neutralizing antibodies, but now we know of a way to actually induce them.”
The only problem is that on subsequent encounters with the virus or a vaccine, the body doesn’t generate those same super-effective antibodies. Instead, for reasons that are unclear, the immune system tends to target newer variations on the virus. That may be effective at the time, but isn’t very helpful down the road when another, slightly different version of influenza comes along.
“When people encounter that virus a second or third time, their antibody response is pretty much completely dominated by antibodies against those more variable parts of the virus,” Guthmiller said. “So that’s the uphill battle that we continue to face with this.”
The trick to getting around this is to design a vaccine that recreates that initial encounter with H1N1, using a version of the HA protein that keeps the conserved, powerful antibody-inducing components, and replaces the variable parts with other molecules that won’t distract the immune system.
“Structural studies were essential to delineate the conserved areas on the HA protein,” said Julianna Han, PhD, postdoctoral research in the Ward lab at Scripps Research Institute and co-first author of the new study. “Now these data can be used to fine-tune vaccine targets.”
In roughly the past century, two of the four flu pandemics have been caused by H1N1 influenza, including the 1918 Spanish flu pandemic that killed as many as 100 million people. Yet the findings of this study are reassuring in the fight against possible future pandemics caused by other H1 viruses.
“The odds of there being another pandemic within our lifetime caused by an H1 virus is quite high,” Guthmiller said. “Just knowing that we actually have the immune toolkit ready to protect ourselves is encouraging. Now it’s just a matter of getting the right vaccine to do that.”