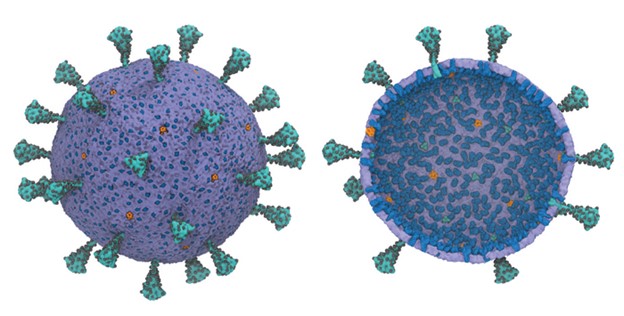
Multiscale model of SARS-CoV-2; exterior (L) and interior (R) views show the spike protein trimers (teal), glycosylation sites (black), membrane proteins (blue), and pentameric envelope ion channels (orange). Credit: Gregory Voth, Univ. of Chicago.
The Texas Advanced Computing Center has announced completion of what it said is the first complete, multiscale model of the SARS-CoV-2 virus, a project that leveraged HPC resources at TACC, the Pittsburgh Supercomputing Center (PSC) and the University of Chicago Research Computing Center (UCRCC).
According to an article published late yesterday by TACC Technical Writer Jorge Salazar, the multiscale model of the virus was developed using TACC’s Frontera system, a Dell system powered by Intel Xeon Platinum processors and currently ranked 9th on the Top500 list of the world’s most powerful supercomputers, along with the Anton 2 systems installed in 2016 at PSC and UCRCC’s Midway2, which also came online in 2016.
“We wanted to understand how SARS-CoV-2 works holistically as a whole particle,” said Gregory Voth, the Haig P. Papazian Distinguished Service Professor at the University of Chicago. Voth is the corresponding author of the study that developed the virus model, published November 2020 in the Biophysical Journal.
TACC said Voth and his team used the all-atom dynamical information on the open and closed states of the virus’s spike protein generated by the Amaro Lab on the Frontera supercomputer, as well as other data. The group initially used the Midway2 cluster to develop the coarse-grained model. The membrane and envelope protein all-atom simulations were generated on the Anton 2 system, a special-purpose supercomputer for molecular dynamics simulations developed and provided without cost by D. E. Shaw Research.
Voth said Frontera has been particularly useful in providing the molecular dynamics data at the atomistic level for feeding into the model.

Frontera supercomputer at TACC
“Frontera has shown how important it is for these studies of the virus, at multiple scales. It was critical at the atomic level to understand the underlying dynamics of the spike with all of its atoms,” he said. “There’s still a lot to learn there. But now this information can be used a second time to develop new methods that allow us to go out longer and farther, like the coarse-graining method.”
It’s hoped the model will help scientists better understand how the virus fuses and enters the host cell; how it assembles itself; and how it buds off the host cell. Early results of the study show how the spike proteins on the surface of the virus move cooperatively.
The full article can be found here.



