Targeting COVID-19 with innovative computational design methods
Yale scientists have developed a new class of antiviral agents that shows promise for creating COVID-19 therapeutics—exhibiting particular effectiveness when used in tandem with the drug remdesivir, another antiviral medication approved for use against the virus.
The new compounds are the first potent, drug-like, non-covalent, and non-peptidic inhibitors of the main protease, or enzyme, in SARS-CoV-2 that is crucial for processing essential viral proteins. A study demonstrating the compounds appears in the journal ACS Central Science.
The work is the latest collaboration of Yale researchers William Jorgensen and Karen Anderson, senior co-authors of the study, who use high-level computation and structural biology methods to design compounds that target viral proteins.
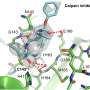
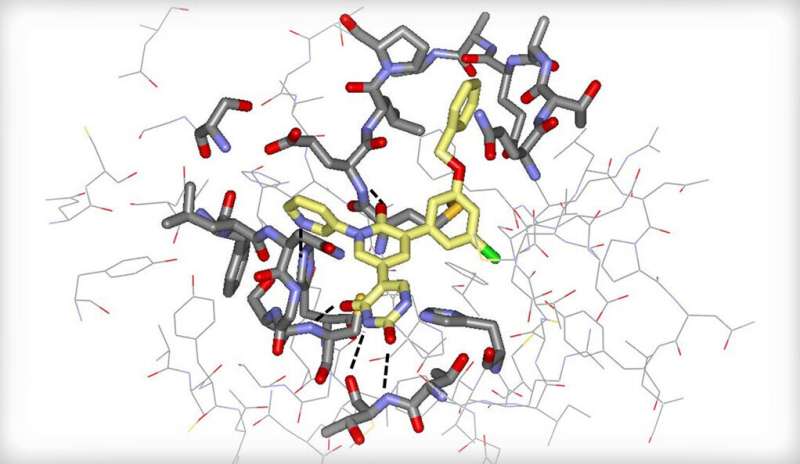
The researchers used in silico screening, performed via computer simulation, to identify an existing, FDA-approved drug—perampanel, an anti-seizure medicine—as a starting point. They employed a state-of-the-art computational technique called free-energy perturbation (FEP) calculation to help design the compounds. (FEP is a method for predicting the binding strength of potential small-molecule drugs to a specific protein.) The compounds were then synthesized and shown to inhibit the viral protease. X-ray crystallography of the drug-protease complexes provided important structural insights.
The new compounds may lead to effective treatments for SARS-CoV-2, the researchers said.
"From the standpoint of efficient molecular design, this is a truly striking and remarkable report," said Jorgensen, Sterling Professor of Chemistry at Yale.
"These structures will almost certainly be used by many other investigators for designing additional antiviral agents," said Anderson, a professor of pharmacology and molecular biophysics and biochemistry at Yale, and co-director of developmental therapeutics for the Yale Cancer Center.
The researchers found that the new compounds showed antiviral activity in cells that have been infected with SARS-CoV-2, and that a representative compound from the study worked especially well with the antiviral drug remdesivir when administered together.
The research team noted the crucial importance of creating new and more effective therapies against COVID-19, its variants, and new coronaviruses, alongside vaccine efforts.
"It should be realized that people will be newly infected by SARS-CoV-2 for years," Jorgensen said. "This includes people who will not take, do not have access to, do not have adequate immunological response, or cannot tolerate the vaccines. There will continue to be great need for small-molecule therapeutics to treat SARS-CoV-2 infections."
The new compounds also may prove particularly useful against future coronaviruses because they work against the main protease, or enzyme, found in many coronaviruses. This is in contrast to the current COVID-19 vaccines, which act upon the highly variable coronavirus spike protein.
"To our knowledge, the only small-molecule inhibitor of the SARS-CoV-2 main protease currently in clinical trials is a repurposed compound that is only administered by IV injection," Anderson said. "Our compound series should have no problem yielding drugs that may be formulated for oral delivery or other forms of delivery such as a nasal spray."
More information: Chun-Hui Zhang et al. Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations, ACS Central Science (2021). DOI: 10.1021/acscentsci.1c00039
Journal information: ACS Central Science
Provided by Yale University