In a healthy human adult, bacterial cells outnumber human cells, but the identity and degree of diversity of these bacteria in a single individual, their variability from person to person, and their role in disease and disease susceptibility has been largely unknown. The goal of the Human Microbiome Project is to characterize the human microbiome and analyze its role in human health and disease.
The human microbiome is defined as the collection of microbes - bacteria, viruses, and single-cell eukaryotes - that inhabits the human body. Microbes in a healthy human adult are estimated to outnumber human cells by a ratio of ten to one, and the total number of genes in the microbiome exceeds the number of genes in the human genome by a factor of at least 200. Even though microbial cells are only one-tenth to one-hundredth the size of a human cell, they may account for up to five pounds of adult body weight.
Although bacteria are often associated with infections, the bacteria that colonize the surface and insides of our bodies are essential for life. We are dependent on these bacteria to help digest our food, produce certain vitamins, regulate our immune system, and keep us healthy by protecting us against disease-causing bacteria.
The composition of the entire collection of microbes that make up the microbiome and its influence on our health and susceptibility to disease is not easily investigated. To date, only a small percentage of the bacteria that comprise the human microbiome have been identified, and a limited number of individual microorganisms have been studied. It simply has not been possible to isolate the vast majority (>95%) of microorganisms and culture them, presumably because the required growth conditions have not or cannot be reproduced in the laboratory. However, recent technological advances in DNA sequencing and the development of a method known as metagenomics have now made it feasible to analyze the entire human microbiome.
Metagenomics
Metagenomics is a sequence-based approach that allows the genetic material from the complete collection of microbes to be analyzed without needing to cultivate the microorganisms. Microbial communities can be harvested from their natural environments, and their DNA sequences can be determined.
The metagenomic approach allows for the identification of microorganisms that were previously unrecognized and gives vastly more information than the analysis of singly isolated microbes. Researchers can determine the relative abundance of the different species and discover which metabolic pathways are encoded by the organisms to gain information about their functions in the body.
Human Microbiome Project

The Human Microbiome Project was launched by the National Institutes of Health in 2007 with the mission to generate the resources and expertise needed to characterize the human microbiome and analyze its role in health and disease. The NIH approved a budget of $170 million for this project over five years, providing support for a number of centers and institutes around the United States, including Baylor College of Medicine. The HMP serves as a "road map" for discovering the roles these microorganisms play in human health, nutrition, immunity, and disease in diverse niches of the human body. A major goal of the HMP is the metagenomic characterization of microbial communities from 300 healthy individuals over time. The HMP is focused on studying the microbes residing in five body areas: skin, mouth, nose, colon and vagina.
Importance of the Human Microbiome Project
The human microbiome makes up about one to two percent of the body mass of an adult. It has been likened to a body organ. But, unlike say a heart or a liver, the importance and function of the microbiome is just starting to be appreciated.
It has long been known that bacteria are involved in certain body processes, such as digesting food and producing vitamins, but the microbiome appears have a much broader impact on our health than was previously realized. The community of microbes in an individual may influence the susceptibility to certain infectious diseases, as well as contribute to disorders such as obesity and diabetes. It may also contribute to the development of some chronic illnesses of the gastrointestinal system such as Crohn's disease and irritable bowel syndrome. Some collections of microbes can determine how one responds to a particular drug treatment. The microbiome of the mother may even affect the health of her children.
A more complete understanding of the diversity of microbes that make up the human microbiome could lead to novel therapies. For example, it may be possible to treat a bacterial infection caused by a "bad" bacterial species by promoting the growth of the "good" bacteria. Microbiome transplants are already being used to combat certain illnesses, such as Clostridium difficile infections, to establish more healthful bacterial populations.
How is the Human Microbiome Project Being Conducted?
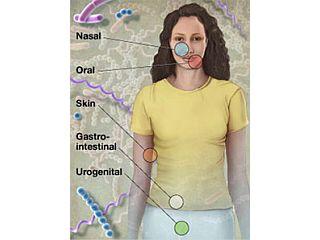
The HMP is a coordinated effort being conducted at 80 institutes across the United States. Baylor College of Medicine is the only site where all aspects of the HMP, from human sampling to sequencing and data analysis, were performed. Initial efforts focused on technological issues involving the development of resources and procedures to accomplish the task of generating and analyzing vast amounts of data.
Because a goal of the HMP is to define a healthy human microbiome, a major challenge was the identification and selection of 300 “normal” subjects. The healthy adult volunteers that researchers recruited for this project were not obese, not on medications, and did not have any chronic health problems or diseases; even minor gum disease was enough to exclude a subject from the study. The clinicians collected multiple samples from different areas of the skin, mouth, nose, stool, and vagina of the volunteers (for a total of 15 sample sites from men and 18 from women) and sampled subjects up to three separate times over the course of a little over one year in an effort to access stability and diversity of the microbiome over time.
Over 11,000 human specimens were obtained. Scientists then purified and sequenced the DNA from them and used information from the bacterially-encoded 16S ribosomal RNA gene to identify and quantify the relative abundance of the bacteria in each sample. Subsequently, additional whole genome sequencing was performed on about 800 of the samples to learn about the genes that encode metabolic functions provided by the microbial communities residing at different body sites. To date, the HMP has generated 3.5 terabytes - or 3.5 trillion bytes - of data, or more than 1000 times the amount produced by the original Human Genome Project.
MVM Researchers' Contributions

Researchers from the Department of Molecular Virology and Microbiology (MVM) have been instrumental in various aspects of the HMP. MVM faculty members Dr. Joseph Petrosino, Dr. Sarah Highlander, Dr. Wendy Keitel, and Dr. James Versalovic (who holds a primary appointment in the Department of Pathology) were involved with the HMP in the early phases of the project's design. They served on HMP Working Groups that established the criteria used to select "normal" subjects and determine the optimal number of body sites and subjects to sample, as well setting guidelines for ethical considerations in subject recruitment. They also worked on developing and testing protocols for standardized sample collection and processing.
Half of the subjects who participated in the HMP were recruited by Baylor. A large part of this success was due to Dr. Keitel's expertise in volunteer recruitment as a result of her work in directing the Vaccine Research Center at Baylor. She served as a principal investigator of the sampling aspect of the Baylor HMP.
The coordination of the human sampling efforts at BCM and at Washington University, in St. Louis, MO, was led by Dr. Versalovic, director of the Texas Children's Microbiome Center, who helped design the methods of clinical sampling. He also directs studies examining the role of the microbiome on healthy children, as well as on children with short bowel syndrome, pediatric irritable bowel syndrome, and recurrent abdominal pain.
Co-principal investigator of the BCM HMP, Dr. Highlander, developed mock communities of bacterial cells and bacterial DNAs in order to test and refine the sequencing and bioinformatics methods later used to characterize the human samples. She also was involved in generating nearly a quarter of the 800 bacterial reference genome sequences that formed a database to map sequences obtained from human samples. She serves as the BCM representative to the International Human Microbiome Consortium.
Dr. Joseph Petrosino, co-principal investigator of the project, is responsible for coordinating the metagenomic and microbiome research and development efforts across BCM and the Texas Medical Center. He serves as the director of the Alkek Center for Metagenomics and Microbiome Research (CMMR).
Alkek Center for Metagenomics and Microbiome Research
The Alkek Center for Metagenomics and Microbiome Research (CMMR) at Baylor, based in the Department of Molecular Virology and Microbiology, serves as an international hub for microbiome research including clinical and basic science applications and advanced bioinformatics analyses. The CMMR was established in 2011 and is directed by MVM faculty member Dr. Joseph F. Petrosino, a nationally recognized leader in metagenomic research. It was founded as an extension to Baylor's involvement in the Human Microbiome Project and is supported in part by a generous donation from the Albert and Margaret Alkek Foundation.
The CMMR builds on the microbiology and virology expertise in the department and collaborates with the Human Genome Sequencing Center, headed by Dr. Richard Gibbs, and the Texas Children's Microbiome Center for pediatric studies under the direction of Dr. James Versalovic.
CMMR researchers are developing molecular and informatics tools and resources to advance diverse clinical and basic research projects pertaining to the organisms that comprise the microbiome, the genetic makeup of these microbes, how these microorganisms interact with human cells and tissues during the course of life and their impact on health and disease. The CMMR provides metagenomic, informatics, model system and molecular biology support and guidance to other researchers and clinical collaborators engaging in these areas of study.
Important Insights
The work on the HMP has yielded fascinating and important insights into the human microbiome. In 2011, the HMP published a report in the journal Science describing sequencing of the first 178 of an anticipated 3000 microbial reference strains, which will serve as a resource for metagenomics studies. Recently, two major papers describing results from the first 242 healthy adults were published in the journal Nature, along with a number of additional publications in PLoS One and other journals. The reports indicate that there is a much greater diversity - both from person to person and between different sites within an individual - than previously realized.
A key finding is the extensive variation in microbiome composition from person to person, even in healthy individuals. There is not a single "normal" or core microbiome; everyone has a personalized microbiome. Nevertheless, different bacterial species may be doing similar jobs in different people. For example, the metabolic processes required to digest complex carbohydrates in the gut may be performed by different bacteria in different individuals.
Notably, researchers observed that most communities of microbes are distinct from one another (such as those on the skin, in the intestine, mouth, and vagina) and do not appear to mix, and not every body site contained members of all the major groups, or phyla, of bacteria known to colonize the human body. Rather, specific groups of microorganisms colonize distinct anatomical niches. Each body site showed a few core or "signature" bacteria with characteristic genes linked to that site, although the relative abundances of these bacteria varied from person to person.
The collections of microbes within different body regions show a surprising degree of diversity. Oral and stool samples had the highest numbers of different types of organisms, followed by the superficial skin samples. The vagina had the least bacterial diversity. There were also substantial differences in the diversity and composition of microbial communities between samples taken from different sites within the same body region, for example, from different areas of the skin.
When the researchers examined which microbes were present, they did not find genes commonly associated with highly pathogenic bacteria, but they did find organisms that are considered opportunistic - ones that can cause disease under certain circumstances. For example, they found Staphylococcus aureus in the noses of 30 percent of the subjects and Escherichia coli in the stools of 15 percent. The absence of disease-causing organisms from the microbiome suggests that people acquire these pathogens from other sources.
Future Goals
The research thus far has been focused on defining a "normal" microbiome. Once the microbiomes from healthy individuals are catalogued, investigators would like to understand the roles these microbes play in our lives and how these communities are impacted by various environmental and genetic factors such as age, geographic location, diet, and ethnicity. They will want to examine the microbiomes of people with various illnesses to determine how the microbiome is altered in different disease states.
The total microbial gene content, or "pan-genome", of about 800 human samples has already been determined and hundreds more are currently being analyzed. Genome sequencing of individual bacteria, viruses and small eukaryotes is continuing to populate the reference "catalog". In addition, studies are underway to examine the viruses and eukaryotes that contribute to the overall microbiome.
Many questions remain. Among them, researchers would like to know how a specific microbiome is established in an individual and how it may change over time, how the human host and microbe community interact, how a particular microbiome affects nutrition and how changes in diet can affect the microbiome, how the microbiome affects immunity and can cause disease, how the microbiome is affected by antibiotics and how the microbiome affects the response to various drugs, and how a microbiome can be altered to improve health.
It is hoped that this research will lead to the development of new treatments and diagnostics for a variety of genetic and infectious diseases.







 Credit
Credit