Cell line development is preparing to let go of its conventional, labor-intensive ways so that it may embrace increasingly powerful and precise optimization methods. The impending change will affect many expression systems, including mammalian cell–based systems, chiefly Chinese hamster ovary select CHO) systems, which have generated more than half of the biopharmaceuticals approved by the Food and Drug Administration over the past decade.
Although CHO systems have been and will remain central to the manufacture of recombinant proteins, they are not as flexible or capable as they could be. To improve CHO and other expression systems, investigators are applying genomic insights to cell engineering, introducing more sophisticated technologies for glycoengineering and pathway delineation, and building machine learning frameworks to realize digital cell culture.
Innovative approaches to cell line development were discussed at the 11th Annual Bioprocessing Summit, which was held recently in Boston. In particular, one of the conference tracks select “Optimizing Cell Line Development”) covered a number of promising developments: the construction of a DNA integration platform containing multiple “landing pad” recombination sites; the engineering of synthetic gene circuits to improve N-glycosylation; the generation of genome/phenome predictions to overcome harmful genome instability; the creation of a “tuning switch” for inducible expression; and the application of nanofluidic technology to advance digital cell culture.
Genomic shuttle sticks the landing
The space shuttle would reach one of its preselected landing sites only after executing a series of well-planned maneuvers, including a brief deorbit burn, delicate attitude adjustments during reentry, momentum-shedding turns in the atmosphere, a glide toward the runway, and then…touchdown. Finally, a drag chute would deploy, helping bring the shuttle to a stop.
Detailed procedures for targeted landings are created by people other than NASA scientists. For example, synthetic biologists at Massachusetts Institute of Technology select MIT) have devised a procedure that allows multiple transgenes to be integrated at precise genomic loci—“landing pads”—with predictable levels of gene expression.
Last year, MIT professor of biological engineering Ron Weiss, PhD, and colleagues unveiled a multiple-landing-pad DNA integration platform. At the Bioprocessing Summit, Weiss elaborated on the group’s work.
“Traditional cell line engineering methods rely on the labor-intensive strategy of random gene integration and high-throughput colony screenings,” he said. “We developed a recombinase-based, site-specific gene integration platform and identified 21 novel sites in the genome of CHO cells that would stabilize long-term expression of transgenes.”
The scientists constructed CHO cell lines containing one, two, or three landing pads at specific loci. Each landing pad contained a recombination site and a selectable marker. Subsequently, the scientists employed a matching recombinase for integration of the DNA payload.
“To validate the functionality and capability for biomanufacturing of these CHO cell lines, we used this technology to precisely integrate up to nine copies of a monoclonal antibody select mAb) representing about 100 kilobases of heterologous DNA,” Weiss noted. “We found a linear increase in the amount of mAb expressed that remained stable for months. The entire process of development and validation took about three years.”
According to Weiss, there are many applications for this technology including biomanufacturing, organoid construction, and human therapy: “Landing pad cells could allow high-throughput construction and precise characterization of synthetic gene networks that dramatically change cells’ phenotype and behavior. It’s my lifelong aspiration and goal to utilize synthetic biology for creating new gene and engineered cell therapies that can treat human diseases. I expect to see initial clinical trials within several years, not decades.”
Bullseye for N-glycosylation
Another MIT investigator, research scientist William C.W. Chen, MD, PhD, spoke at the Bioprocessing Summit. He said that synthetic biology and cell engineering can also provide new toolkits for N-glycosylation in recombinant protein production.
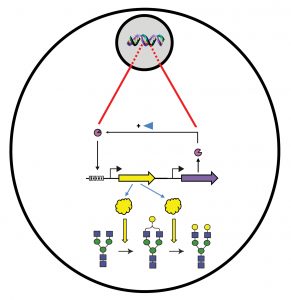
At MIT, William C.W. Chen, MD, PhD, and colleagues have developed a genomically integrated synthetic biology toolkit to control cellular N-glycosylation. By using the toolkit to realize recombinase-based, site-specific genomic integration of small molecule-inducible gene circuits, researchers at MIT’s Synthetic Biology Center can precisely control N-glycosylation of monoclonal antibodies expressed in CHO cells.
Reliable N-glycosylation is important for the wide variety of mAb therapeutics as it impacts safety, efficacy, stability, and pharmacokinetics,” Chen emphasized. “Unfortunately, we lack the necessary toolkits for ensuring precise control of N-glycosylation during mAb production.”
Important mAb properties such as stability, solubility, appropriate folding, and biological activity of mAb therapeutics derive largely from an N-glycosylation site in the Fc region of immunoglobulin molecules. Unfortunately, during production, the high number of different sugar moieties and their complex linkages can form different glycan structures and thus yield varied biological forms and functions.
Chen indicated that colleagues at MIT and Pfizer select under the direction of MIT colleagues Weiss, Timothy K. Lu, MD, PhD, and Douglas A. Lauffenburger, PhD) are engineering CHO cells with synthetic gene circuits to fine-tune N-glycosylation of stably expressed mAbs. To accomplish the feat, they knocked out two key glycosyltransferase genes and then replaced them with synthetic versions of the same genes that were under the control of constitutive or inducible synthetic promoters.
“This strategy enabled the precise control of both genes and tunable levels of fucosylated or galactosylated mAb,” Chen declared. “The technology may also provide precise and comprehensive control of other recombinant proteins.”
Although most applications utilize CHO cells, Chen noted that these are not the only cells of interest for synthetic biology toolkits: “We also want to test other cell types to see if these platforms provide a universal or cell-specific approach.”
Solving CHO genome instability
Despite being the most common mammalian cell line used to produce therapeutic proteins, CHO cells have a notoriously unstable genome, asserted Derrick C. Scott, PhD, assistant professor of biological sciences and director of bioinformatics at Delaware State University: “The cells you start with are often different than the cells you end up with due to genome shifting.
“This side effect can cause those cells to be discarded, which has the end result of driving up the cost of medications. Currently, the annual cost for a person using CHO-based biologics can exceed $100,000.”
Scott and colleagues are endeavoring to determine which genes are driving this instability. “We have begun a four-pronged approach,” he reported, “to understand the relationship between the CHO genome and the physical characteristics of the expressed protein, which is the phenome.”
Initially, collaborators from Clemson University, led by principal investigator Sarah Harcum PhD, professor of bioengineering, grow the CHO cells while varying the growing conditions—for example, altering the temperature or pH or adding chemicals—to induce instability. Next, they sequence the RNA of the samples. This information is channeled to Scott, who uses a battery of bioinformatics tools to compare the CHO reference genome with the challenged CHO genomes.
According to Scott, “This allows us to pinpoint transcriptional changes in genes in response to the growth conditions. Next, we send this data to Nicholas Sandoval, PhD, assistant professor of chemical and biomolecular engineering, Tulane University, who will expand our knowledge by performing knock-out or knock-in studies. Finally, all this data will be funneled to Kelvin Lee, PhD, professor of chemical and biomolecular engineering, University of Delaware, who is the science lead of the project.”
The project is already well underway and generating massive amounts of data. “Our team is well versed in CHO biology, and we have completed preliminary studies,” Scott continued. “We are primed for the end game and expect to determine the best targets to ultimately create a more stable CHO cell line that will decrease manufacturing costs and increase patient access to expensive medications.”
Cumate gene switch
Isolation of stable high-expressing CHO cell lines from heterogeneous populations is a daunting task that typically requires six to nine months of intensive investigations, said Yves Durocher, PhD, principal research officer, Human Health Therapeutics Research Center, National Research Council Canada.
“The low frequency of high-producing clones,” he pointed out, “is a reflection of three major stresses cells incur during the process. First, the transfection results in reduced viability. Second, the stringent selection process imposes a strong metabolic pressure. Third, typical constitutive expression systems force cells to continuously generate high levels of product that induces a metabolic burden and endoplasmic reticulum stress. This may result in low-expressers outgrowing high-expressing cells.”
Durocher and his team approached expression in a way that cooperated with the cells’ biology to reduce one of these stresses. “We created a ‘tuning switch’ for inducible expression,” he explained. “Initially, we developed a CHO cell line that harbored components of the cumate-inducible system. For generating stable pools, we optimized transfection and selection using plasmid vectors containing genes of interest select antibodies or Fc fusions) controlled by the cumate gene switch promoter. Surprisingly, we found that during pool selection, cells with reduced levels of expression select that is, in the ‘off state’) showed up to threefold increased productivity in a fed-batch process as compared to pools selected while continuously expressing the gene of interest select ‘on state’).”
Not only did inducible pools outperform those made with constitutive promoters, even difficult-to-express proteins such as cytokines and G protein–coupled receptors could be reliably, stably, and quickly expressed. “We also determined that 70% of clones were stable even after 55 generations of continuous culture,” Durocher reported. “Further, cumate has no negative impact on CHO cells. The entire process from transfection to a 10-L pool harvest takes about four to five weeks, resulting in greatly reduced cost and development time.”
The cumate-inducible clones derived from those pools are being used for clinical trials and commercial production. One biosimilar product is currently being marketed in Asia, while three products are underdoing Phase I/II clinical trials in the United States and Europe and a fourth will soon be in a Phase I trial.
Digital cell culture
“Successful development of a therapeutic biologic involves a well-orchestrated and resource-intensive process to isolate a master cell bank that will live for the lifetime of the product,” commented Jennitte Stevens, PhD, executive director, process development, drug substance technologies, Amgen. However, traditional methods to produce the master cell are relatively inefficient and slow.
“Often researchers are manipulating and analyzing thousands of wells on multiple plates, followed by assessing cell growth and productivity on disconnected endpoint analytical equipment,” she continued. “Even though automation helps, what is lacking is a modular framework that can unify workflows and enhance overall efficiency of the process.”
To overcome these bottlenecks, Amgen is shifting toward a “digital cell culture” paradigm. Stevens explained, “Digital cell culture is the intersection of cell culture, automation, and data analytics, and it will provide scientists a faster and less resource-intensive way to run standard types of cell culture experiments while obtaining more data.”
To advance this strategy, Amgen has invested in a software-driven nanofluidic cell culture and analytic technology, developed by Berkeley Lights, which utilizes light to activate photosensitive switches for isolating and positioning individual cells on a chip. Thousands of cells flow into the nanofluidic channels and are gently guided by light beams into individual tiny pens. The chip and its integrated technologies serve as a miniscule laboratory that can run a range of experiments on individual single cells at a scale 50,000 times smaller than traditional methods.
“We can quickly determine which cells are making high levels of therapeutic protein such as antibodies, and also obtain data on important cellular properties at a single-cell level,” reported Stevens. An entire chip image can be processed in about eight minutes, and data required to analyze and select highly productive cell lines are generated in hours or days rather than weeks or months.
A second Amgen strategy incorporates advanced machine learning into the digital cell culture process to enhance the predictive power for selecting the key cell lines. According to Stevens, “We can utilize this information to gain insights for defining population characteristics from production cell lines. This is not possible with traditional analytic techniques.”
Outsourcing to Enhance Efficiency
Early-stage biotech companies are often so focused on collecting material as quickly and cost-effectively as possible that they may overlook potential stumbling blocks that could cost time and money. Outsourcing could enhance efficiency.
MilliporeSigma reports that it has designed an adaptive solution to provide proof-of-concept or a stable cell line. According to Sebastien Ribault, PhD, senior director, Global BioReliance® End-to-End Delivery and Sales, “The regular or fast-track custom service is modeled to the needs of small and emerging biotechs from DNA, through minipools and clone selection, to process development manufacturing and, finally, the clinic. Projects are assigned to a dedicated project manager and a technical leader who provide risk assessment and regulatory expertise to fully document monoclonality and stability validation based on regulatory expectations and a statistical approach. There are no royalty fees, and customers have the freedom to transfer technology at any stage, to any partner.”


