This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
A new control switch could make RNA therapies easier to program
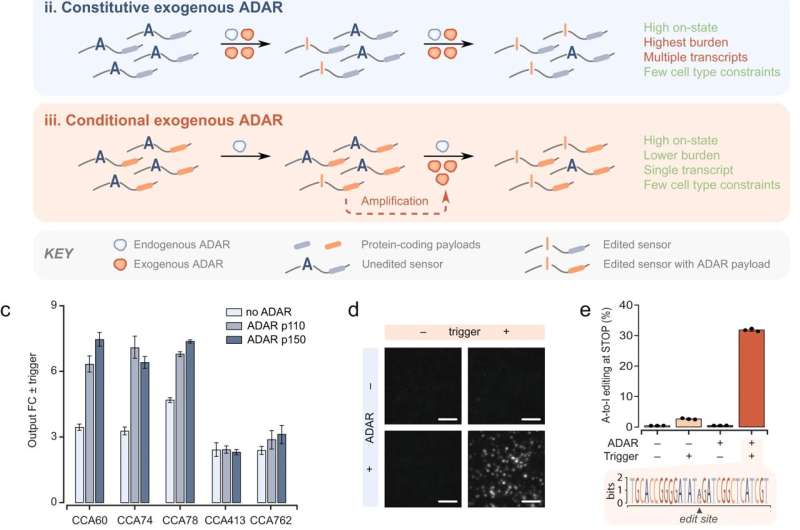
Using an RNA sensor, MIT engineers have designed a new way to trigger cells to turn on a synthetic gene. Their approach could make it possible to create targeted therapies for cancer and other diseases, by ensuring that synthetic genes are activated only in specific cells.
The researchers demonstrated that their sensor could accurately identify cells expressing a mutated version of the p53 gene, which drives cancer development, and turn on a gene encoding a fluorescent protein only within those cells. In future work, they plan to develop sensors that would trigger production of cell-killing proteins in cancer cells, while sparing healthy cells.
"There's growing interest in reducing off-target effects for therapeutics," says James Collins, the Termeer Professor of Medical Engineering and Science in MIT's Institute for Medical Engineering and Science (IMES) and Department of Biological Engineering. "With this system, we could target very specific disease cells and tissues, which opens up the possibility of identifying cancer cells and then delivering highly potent therapeutics."
This approach could also be used to develop treatments for other diseases, including viral or bacterial infections, the researchers say.
Collins is the senior author of the new study, which appears in Nature Communications. The lead authors of the paper are MIT postdocs Raphaël Gayet Ph.D. '22 and Katherine Ilia Ph.D. '23, senior postdoc Shiva Razavi, and former postdoc Nathaniel Tippens.
An RNA control switch
Many experimental therapies involving DNA or RNA delivery—such as gene therapy, CRISPR-based therapies, and RNA interference—are currently under development. An important aspect of such therapies is making sure they are turned on only in the target cells, using a programmable control switch.
In 2021, Collins' lab developed a control switch for RNA therapies known as eToehold. This system is based on RNA molecules called internal ribosome entry sites (IRES), which can be designed to respond to a particular messenger RNA (mRNA) sequence within a cell. However, these systems are difficult to design because their function depends not only on the sequence of the IRES molecule, but also its three-dimensional structure.
For the new study, the researchers wanted to create a system that would be easier to program. Instead of IRESes, they decided to use a synthetic strand of RNA, also called an RNA construct, as the targeting molecule. This would allow them to reprogram the construct to target different mRNA molecules, by simply changing the RNA sequence of the construct.
"With this new system, we have a very straightforward, programmable way of creating control elements that will respond only in the presence of those target sequences," Collins says.
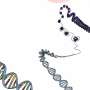
To achieve that, the researchers harnessed an enzyme that naturally exists in most animal cells, known as adenosine deaminase acting on RNA (ADAR). This enzyme performs base editing of RNA molecules, converting mismatched adenosine bases to inosine. This helps cells to fend off invading viruses, among other functions.
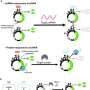
ADAR can detect and repair mismatches in double-stranded RNA, so the researchers designed their sensor RNA construct so that contains a sequence complementary to their target mRNA but with one mismatch. This draws the attention of ADAR that naturally exists in the cell, which repairs the mismatch.
When ADAR converts adenosine to inosine in the RNA sensor, that edit removes a stop codon in the sequence. After this stop codon is removed, the cell begins reading the RNA construct, which the researchers designed to contain two protein-coding genes. The first is for a reporter molecule—in this case, a fluorescent protein that allows the researchers to see that the synthetic gene was activated. In future versions, this could be replaced by a gene encoding a therapeutic agent.
The other synthetic gene encodes a stripped-down version of the ADAR enzyme. As more ADAR is produced, the enzyme finds and activates more copies of the synthetic RNA construct. This creates a positive feedback loop that enhances the expression of the fluorescent reporter gene.
Other researchers have shown that ADAR can be used for this kind of RNA targeting, but most of those studies were limited to cells that naturally produce larger amounts of the enzyme, such as neurons.
"We only require a very small amount of ADAR to initially trigger the network. And then through a positive feedback design, that small trigger gets the cells to express high levels of a compact form of that enzyme that's built into the construct," Collins says. "This broadens the potential application uses for the system in that now it's not restricted to cells that have large background levels of ADAR."
High precision
In tests in human cells, the researchers explored whether this sensor, which they named DART VADAR (detection and amplification of RNA triggers via ADAR), could distinguish between very similar mRNA sequences. To do that, they inserted the sensor construct into human cells that had either the normal version of the p53 gene or a mutated version, which differs by only a single base pair and is known to drive cancer development.
"We show that you can get very high resolution and very high precision for these sensors," Gayet says. "With a carefully designed sensor, you can get a different level of activation depending on whether or not the cells produce some RNA that includes a mutation."
In another set of experiments in mouse cells, the researchers showed that the sensor construct could distinguish between closely related cell types that differentiate into either bone or muscle cells.
Because the researchers used a trimmed down version of the ADAR enzyme, which is only about 1,600 base pairs, the entire construct can easily fit in an AAV vector—a type of modified, harmless virus that is often used in the clinic to deliver genetic material in humans.
The researchers now plan to try testing their system in animal models of cancer, to see if they can deliver synthetic constructs that would selectively kill tumor cells by producing lethal compounds only within those cells.
More information: Raphaël V. Gayet et al, Autocatalytic base editing for RNA-responsive translational control, Nature Communications (2023). DOI: 10.1038/s41467-023-36851-z
Journal information: Nature Communications
Provided by Massachusetts Institute of Technology
This story is republished courtesy of MIT News (web.mit.edu/newsoffice/), a popular site that covers news about MIT research, innovation and teaching.