By MaryAnn Labant
Back in 2016, a group of researchers led by Harvard’s David Liu, PhD, published a paper in Nature that reported the development of base editing. Base editing, they wrote, is “a new approach to genome editing that enables the direct, irreversible conversion of one target DNA base into another in a programmable manner, without requiring double-stranded DNA backbone cleavage or a donor template.” They added that base editing could reduce the risk of off-target mutations and had the potential to “efficiently correct a variety of point mutations relevant to human disease.”
Similar work at the time was accomplished by Keiji Nishda, PhD, and his colleagues at Kobe University. Many additional base editing systems have been developed by academic and commercial researchers in the years since. And that’s not all. In 2019, yet another genome editing system capable of avoiding double-stranded DNA breaks was introduced. This system is called prime editing, and it was developed, not so coincidentally, by researchers led by Liu.
“[Prime editing is] a versatile and precise genome editing method that directly writes new genetic information into a specified DNA site using a catalytically impaired Cas9 endonuclease fused to an engineered reverse transcriptase, programmed with a prime editing guide RNA (pegRNA) that both specifies the target site and encodes the desired edit,” the researchers wrote in Nature. “Prime editing … has complementary strengths and weaknesses compared to base editing and induces much lower off-target editing than Cas9 nuclease at known Cas9 off-target sites. Prime editing substantially expands the scope and capabilities of genome editing, and in principle could correct up to 89% of known genetic variants associated with human diseases.”
Today, base editing and prime editing systems for clinical applications are in development. Several such systems are highlighted in this article. Although these systems are cutting edge—or rather nicking edge—they still need to attend to familiar challenges such as packaging and targeted delivery.
Prime editing evolves
“We believe that prime editing is highly specific and precise because the prime editing machinery has to go through three annealing steps to make a genetic change,” says Keith Gottesdiener, MD, president and CEO, Prime Medicine. “It is unlikely that it would land on the wrong spot and unlock all three keys.”
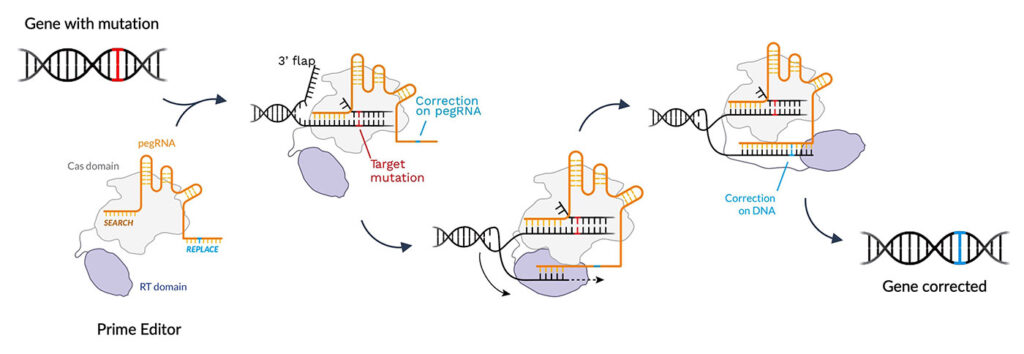
According to Gottesdiener, prime editing can correct the 12 different single-base-pair mismatches, repair small indel mutations, and remove and replace large genomic sequences—all in a programmable way. “In programmable editing, the edit is precisely targeted,” he emphasizes. “We can pick, to a base pair, where we want an edit to occur. And since the machinery runs and is guided by a small gRNA, we can swap that and move to another place in the genome.”
Prime editing works in a vast array of rapidly dividing or quiescent cells. A once-and-done treatment, the DNA is returned to wild type at the normal genetic locus.
“We enhanced and improved the original technology,” Gottesdiener asserts. “It is almost too good to be true.” He cautions, however, that “delivering the technology to any site has challenges similar to those seen by others.” Familiar delivery options include electroporation for ex vivo applications; lipid nanoparticles (LNPs) for targeting the liver; and adeno-associated virus (AAV) vectors for targeting other strategic locations.
Prime Medicine has a broad pipeline. Although such a pipeline gives the company a range of options, it also poses a management challenge. “All indications could advance to the clinic,” Gottesdiener notes. “However, some programs will progress faster, some we will partner out, and others will be pruned.”
In the initial pipeline, there is an emphasis on immediate indications and differentiation programs. Prime selected these indications after balancing considerations such as defined genetics, emerging medical priorities, input from Prime’s thought leader partners, and regulatory and delivery challenges. The differentiation program focuses on unique disease characteristics such as tackling repeat expansions found in Huntington’s disease, a form of amyotrophic lateral sclerosis, and some forms of ataxia. Prime plans to submit an IND for its first indication, chronic granulomatous disease, in 2024.
Base editing enters U.S. clinical trials
Giuseppe “Pino” Ciaramella, PhD, president of Beam Therapeutics, observes that base editing modifies CRISPR-based technology in two critical ways. First, base editing needn’t make double-stranded breaks, which can lead to genomic rearrangements and other unwanted consequences. Second, base editing typically fuses the CRISPR protein to an additional human protein, a deaminase that is capable of chemically converting one nucleobase to another directly on the gene.
A limitation of base editors is that they perform only certain types of single-base-pair edits. That is, they can perform only transition mutations (purine to purine, or pyrimidine to pyrimidine), not transversion mutations (purine to pyrimidine, or pyrimidine to purine). They cannot be used to perform insertions or deletions or replace any stretch of DNA desired. Nonetheless, because they are tools capable of making transition point mutations with high precision and efficiency, base editors have potential in a variety of therapeutic applications.
The lack of double-stranded breaks confers a significant advantage by allowing several simultaneous edits. One system that exploits these simultaneous edits is a Beam platform called Engineered Stem Cell Antibody Paired Evasion (ESCAPE). It combines antibody-based conditioning with multiplex gene edited hematopoietic stem cells.
“We can make an edit aimed at modifying sickle-cell disease as well as an edit that makes the edited cells resistant to a bespoke antibody that can selectively target unedited cells and create a niche in the bone marrow that allows the edited cells to engraft following a transplant,” Ciaramella explains. This may enable a nontoxic conditioning regimen, unlike the current standard-of-care regimen, which can cause sterility and other side effects.
The company uses clinically validated delivery technologies to deliver mRNA encoding the editor. Ex vivo programs use electroporation, and in vivo programs encapsulate the mRNA into LNPs that can target the liver. Novel LNP formulations are being developed to target other tissues like immune cells or stem cells in the bone marrow.
Beam’s lead program for sickle-cell disease, BEAM-101, is the first U.S. clinical base editing program. Ciaramella notes that Beam’s regulatory package was consistent with that required for other ex vivo gene editing programs.
In addition to developing its base editing technology, Beam is collaborating with other companies. Through an agreement with Prime Medicine, Beam has the exclusive right to develop prime editing technology for the creation or correction of any single-base transition mutation, as well as for the treatment of sickle-cell disease. Also, Beam is working with Verve Therapeutics. In this collaboration, Verve has exclusive access to Beam’s base editing, gene editing, and delivery technologies for human therapeutic applications against certain cardiovascular targets. Although Beam also has access to RNA editing technology, the company, Ciaramella says, has not found applications where RNA editing would provide a benefit over DNA editing.
Permanently lowering blood cholesterol
Cardiologist and scientist Sekar Kathiresan, MD, highlights three insights into cardiovascular disease that have emerged from human genetics research. First, if an individual’s blood cholesterol level is low throughout their lifetime, they are very unlikely to develop a heart attack. Second, some individuals resistant to heart disease carry mutations—resistance mutations—that naturally turn off cholesterol-raising genes. Third, three of the resistance mutations affect the low-density lipoprotein (LDL) cholesterol pathway, four of them affect the triglyceride-rich lipoprotein (TRL) cholesterol pathway, and one of them affects the lipoprotein(a) (Lp(a)) cholesterol pathway.
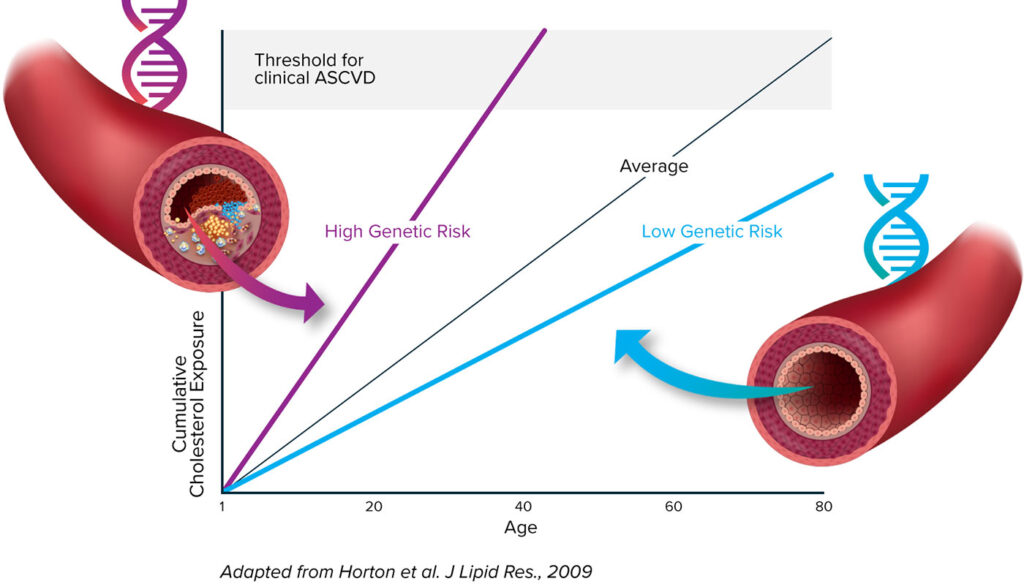
“CRISPR technology allowed us to develop a medicine that would mimic these natural resistance mutations,” Kathiresan continues. By “us,” he means the development team at Verve Therapeutics, the genetic medicines company that he serves as co-founder and CEO. “Our once-and-done gene editing medicines,” he asserts, “are designed to permanently lower blood cholesterol.”
VERVE-101, the company’s most advanced product candidate, targets the PCSK9 gene to lower LDL cholesterol levels. VERVE-201, the company’s next most advanced product candidate, targets the ANGPTL3 gene to lower TRL cholesterol levels. Both the VERVE-101 and VERVE-201 programs rely on base editing technology. To target the LPA gene and lower Lp(a) cholesterol levels, Verve is designing a custom editor.
Delivery benefits from refinement. Standard LNPs are little fat bubbles that carry cargo. In the case of VERVE-101, the cargo is the mRNA for the CRISPR base editor as well as a gRNA that directs the editor to the PCSK9 gene. “We added a ligand, a GalNAc sugar molecule, to a standard LNP,” Kathiresan says. “Since the GalNAc receptor is only expressed on hepatocytes, a GalNAc LNP has the potential to be more potent and specific.”
VERVE-101 is currently in a Phase I trial, called heart-1, to evaluate safety and efficacy in patients with heterozygous familial hypercholesterolemia (HeFH), a genetic form of high cholesterol and premature heart disease. A morbid disease, HeFH is characterized by severely elevated blood levels of LDL cholesterol and premature atherosclerotic cardiovascular disease. Current LDL cholesterol–lowering regimens of daily pills or intermittent injections place a heavy treatment burden on patients, providers, and the healthcare system, and fewer than 5% of HeFH patients globally attain LDL cholesterol goal levels.
The heart-1 trial is currently enrolling in New Zealand and the United Kingdom. Clinical data are expected later this year. The company is working with the FDA to open an IND in the United States.
Engineering nuclease characteristics
Clinical applications of CRISPR-Cas9 technology must demonstrate safety and flexibility, says David Baram, PhD, president and CEO of Emendo Biotherapeutics. Safety, he suggests, is largely a matter of avoiding off-target effects. “CRISPR- Cas9 targets mostly what you want it to cut,” Baram says. “But mostly it is not sufficient for safety in its wild-type form.” With respect to flexibility, he emphasizes the need for multiple solutions: “To be able to treat disease in the most effective way, a portfolio of editing strategies allows targeting of more diseases and loci on specific genes.”
Perhaps the best-known nuclease in gene editing is the Cas9 variant isolated from the bacterium Streptococcus pyogenes (SpCas9). There are thousands of other nucleases, however, some of which are superior in activity, the number of sites they can target, specificity, and packaging efficiency. A panel of nucleases provides the option of finding the best nuclease for treating a specific disease. “Our nucleases are highly active and very precise, and they can target 90% of the human genome,” Baram asserts. “SpCas9 covers less than 10%.”
A team of computational experts and scientists generates the panel of nucleases. The company’s engineering platform then adapts these wild-type nucleases for specific purposes. An example is allele-specific editing of autosomal dominant disease in which there is one healthy allele and one mutated allele. “We can target the mutated allele even if it has only a minor single nucleotide difference,” Baram states. This has opened a window to treating autosomal dominant diseases in a single step.
Emendo’s lead indication is severe congenital neutropenia, a neutrophil maturation disorder that eventually causes an immunodeficiency, making patients highly susceptible to bacterial infections. The standard therapy for the disease includes injections of granulocyte colony-stimulating factor, which can help restore immune system function. To treat and hopefully cure the disease, Emendo has developed a novel CRISPR-based ex vivo gene editing strategy (OMNI A1), one that involves specific excision of a disease-causing mutant allele of the ELANE gene. Emendo plans for it to be in the clinic by the end of 2023.
A familial hypercholesterolemia treatment is also in the pipeline. This treatment involves targeting the LDLR gene (which encodes the LDL receptor) rather than the PCSK9 gene (which encodes a protein that regulates LDL receptor expression) to elevate the level of expression of LDL receptor on the surface of hepatocytes and take LDL cholesterol from the blood into the cells. The editing strategy excises a regulatory element in the LDLR gene that directly influences the expression levels of LDL receptor on the surface of the membrane. Data show that using the approach elevates LDL cellular intake levels threefold more than does knocking out PCSK9 and administering statins combined.
A modular base editing platform
Revvity for Life Sciences’ Pin-point™ Base Editing Platform incorporates the following elements: a gRNA; a nickase (based on Cas9 or a similar protein to avoid creating double-stranded DNA breaks); and a deaminase (or another effector molecule). The system is assembled using an aptamer on the gRNA and an aptamer binding protein on the deaminase or other effector molecule.
What distinguishes the Pin-point Base Editing Platform from other base editing systems? With the Pin-point Base Editing Platform, the deaminase and the nickase are not fused to each other. Different deaminases and nickases can be swapped out independently.
“By using a gRNA without an aptamer, it is possible to target the nicking function of the nCas9 to insert DNA sequences into targeted sites in the genome,” explains Michelle Fraser, PhD, business unit manager, Pin-point Base Editing Platform, Revvity for Life Sciences. “This enables multiplex editing, causing the knockout of genes or the correction of genetic errors at the single- base level and also, concurrently, the insertion of genes.”
The Pin-point Base Editing Platform achieved >75% base conversion from C to T using a cytosine base editor in a multiplex edit of four different immunogenic loci (B2M, CD52, PDCD1, and TRAC) in T cells. All four of these loci were knocked out in over 50% of the edited cell populations.
The Pin-point Base Editing Platform was also used to generate chimeric antigen receptor (CAR) T cells in which these same four immunogenic markers were knocked out and the CD19-targeting CAR was knocked in. A tumor- cell-killing assay confirmed that these CAR T cells were highly effective. Multiplex base editing has also been demonstrated in induced pluripotent stem cells at a similar efficiency.
The Pin-point Base Editing Platform can be licensed from Revvity for Life Sciences. Additionally, a tiled screening service is available, where the Pin-point platform is used to edit every available C residue in a target gene or genes of interest. The phenotypes of edited cells are then used to identify functional domains or characterize suspected genetic variants.
The array of enzymes—Cas variants, nickases, deaminases, and other effectors—is rapidly expanding through enzyme discovery programs that are looking at biodiverse sources, as well as through laboratory-based protein engineering and directed evolution projects. According to Revvity for Life Sciences, the modularity of the Pin-point Base Editing Platform provides a unique opportunity to replace the nickase and/or deaminase with newly developed variants.


