Shortly before his death, Alan Turing published a provocative paper outlining his theory for how complex, irregular patterns emerge in nature—his version of how the leopard got its spots. These so-called Turing patterns have been observed in physics and chemistry, and there is growing evidence that they also occur in biological systems. Now a team of Spanish scientists has managed to tweak E. coli in the laboratory so that the colonies exhibit branching Turing patterns, according to a recent paper published in the journal Synthetic Biology.
"By using synthetic biology, we have a unique opportunity to interrogate biological structures and their generative potential," said co-author Ricard Solé of Universitat Pompeu Fabra in Barcelona, Spain, who is also an external professor at the Santa Fe Institute. "Are the observed mechanisms found in nature to create patterns the only solutions to generate them, or are there alternatives?"
In synthetic biology, scientists typically stitch together long stretches of DNA—which can be taken from other organisms, or be entirely novel—and insert them into an organism's genome.
As we've reported previously, Turing was attempting to understand how natural, nonrandom patterns emerge (like a zebra's stripes or a leopard's spots), and he focused on chemicals known as morphogens in his seminal 1952 paper. He devised a mechanism involving the interaction between an activator chemical that expresses a unique characteristic (like a tiger's stripe) and an inhibitor chemical that periodically kicks in to shut down the activator's expression.
Both activator and inhibitor diffuse throughout a system, much like gas atoms will do in an enclosed box. It's a bit like injecting a drop of black ink into a beaker of water. Normally this would stabilize a system: the water would gradually turn a uniform gray. But if the inhibitor diffuses at a faster rate than the activator, the process is destabilized. That mechanism will produce a so-called "Turing pattern": spots (like on a leopard) or stripes (like on a tiger).
James Murray, emeritus professor of mathematical biology at the University of Oxford and an applied mathematician at Princeton, imagined a field of dry grass dotted with grasshoppers for an article I wrote for Quanta back in 2013:
If the grass were set on fire at several random points and no moisture were present to inhibit the flames, Murray said, the fires would char the entire field. If this scenario played out like a Turing mechanism, however, the heat from the encroaching flames would cause some of the fleeing grasshoppers to sweat, dampening the grass around them and thereby creating periodic unburned spots in the otherwise burned field.
Scientists have tried to apply this basic concept to many different kinds of systems. For instance, neurons in the brain could serve as activators and inhibitors, depending on whether they amplify or dampen the firing of other nearby neurons—possibly the reason why we see certain patterns when we hallucinate. There is evidence for Turing mechanisms at work in zebra-fish stripes, the spacing between hair follicles in mice, feather buds on a bird's skin, the ridges on a mouse's palate, as well as the digits on a mouse's paw. Certain species of Mediterranean ants will pile the dead bodies of ants into structures that seem to exhibit Turing patterns, and there is evidence of Turing patterns in the movement of Azteca ant colonies on coffee farms in Mexico.
In essence, it's a type of symmetry breaking. Any two processes that act as activator and inhibitor will produce periodic patterns and can be modeled using Turing's diffusion function. The challenge is moving from Turing's admittedly simplified model to pinpointing the precise mechanisms serving in the activator and inhibitor roles. This is especially challenging in biology, where scientists are keen to shed more light on the question of how a complex embryo can emerge from tissue that is completely homogenous.
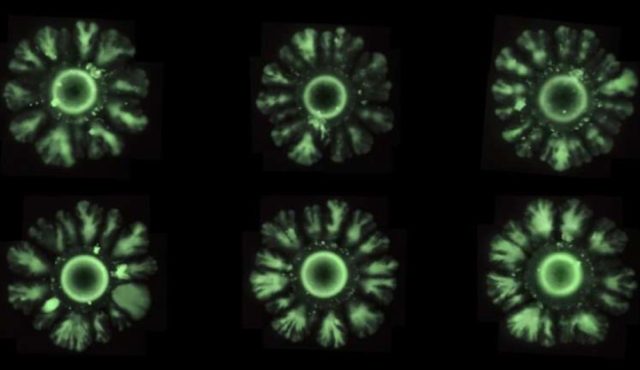
For this latest study, Solé and his collaborators opted to work with colonies of E. coli, genetically engineering the bacteria to introduce a mechanism to generate spatial patterns. "We wanted to build symmetry breaking that is never seen in colonies of E. coli, but is seen in patterns of animals, and then to discover which are the essential ingredients needed to generate these patterns," said co-author Salva Duran-Nebreda, now a postdoc at the Institut de Biologia Evolutiva in Barcelona.
Solé's team found inspiration in the underlying mechanisms of how ants and termites build their nests. Their altered E. coli system consisted of three critical components: a group of regular-sized cells that divided and diffused normally (the activator); a group of elongated cells unable to divide or diffuse (the inhibitor); and a molecule known as a lactone that helps regulate gene expression in E. coli, enabling them to communicate through so-called quorum sensing.
The researchers then observed how the colony grew and evolved. The shape started out as a circle, but as the days progressed and it kept spreading outward, the colony began sprouting regularly spaced "branches" around the edge, in keeping with Turing's theory—like a flower with petals.
"We have seen that by modulating three ingredients we can induce symmetry breaking. In essence, we have altered cell division, adhesion between cells and long-distance communication capacity (quorum sensing), that is to say, perceive when there is a collective decision," said Duran-Nebreda.
The authors hope to apply their findings to other biological systems, such as social insects. Their work provides "a new conceptual framework to create Turing-like patterns in microbial communities, and the relevance of this study goes far beyond this specific implementation," said Solé. "We suspect that hidden under the complexity of entangled gene interactions lies the kind of self-organization principles envisioned by Turing."
DOI: Synthetic Biology, 2021. 10.1021/acssynbio.0c00318 (About DOIs).
Listing image by Ricard Solé
reader comments
31