DNA Nanotechnology: Building Blocks of the Future
Content
DNA nanotechnology is a field of nanotechnology that uses DNA molecules as building blocks to create structures and devices on the nanoscale, including the emerging field of DNA computing.
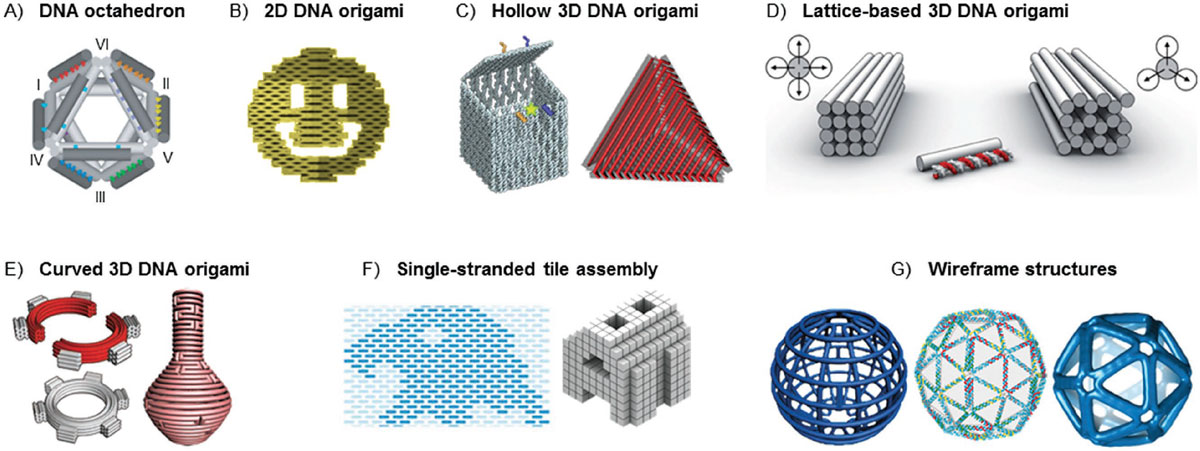
DNA nanotechnology has become an intense research field, with hundreds of research labs engaged in it, based on the idea that DNA can be used as a programmable structural material for the creation of artificial, rationally designed nanostructures.
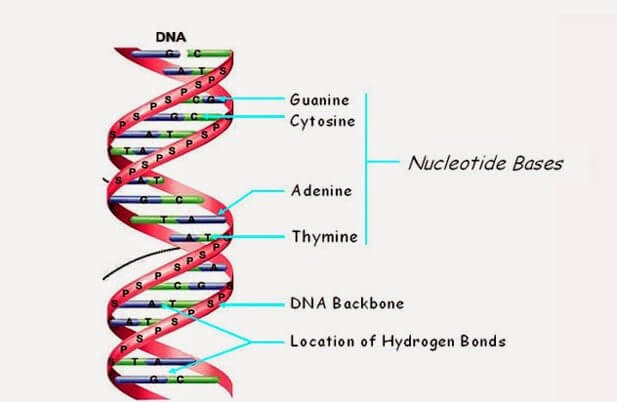
Most people associate DNA with life rather than computers, but it is essentially a four-letter code that transmits information about an organism. DNA is composed of four types of nucleotides, or bases, each designated by a letter: adenine (A), thymine (T), guanine (G), and cytosine (C) – or CGAT for short. These bases intertwine in a spiral pattern to create the molecule's familiar double helix structure. The arrangement of these letters into sequences produces a code that instructs an organism on how to develop. The complete set of DNA molecules constitutes the genome, which serves as the blueprint for an individual's body.
The highly specific nucleotide interaction following the Watson-Crick base pairing rule, as well as the chemically tunable groups present in DNA, makes it possible to encode both structural and chemical/functional information.
DNA is often used as a system for organizing other materials than DNA to somewhat lower resolution than the crystallographic ideal. Gold nanoparticles, proteins, viruses, and other nanoscale species have all been organized, usually using origami.
DNA nanostructures have been utilized in many applications in biology, chemistry, and physics, including electric circuits, DNA-based computing, and nanofabrication.
History of DNA in nanotechnology
In 1982, Nadrian Seeman, a chemist and crystallographer at New York University, laid the conceptual foundation for what is now known as DNA nanotechnology. Seeman's work on the structural properties of DNA led him to realize the potential for using DNA molecules as building blocks for creating nanostructures.
Seeman's pioneering work was based on the idea that the specific base pairing properties of DNA could be exploited to create new, highly ordered structures at the nanoscale. He demonstrated that DNA strands could be designed to self-assemble into complex shapes, such as cubes, tetrahedra, and other polyhedral.
This approach, known as structural DNA nanotechnology, has since been developed into a powerful tool for creating precise and programmable nanostructures.
Another key development in DNA nanotechnology was Paul Rothemund's invention of DNA origami in 2006. It involves using a long single-stranded DNA (ssDNA) that is folded into designated shapes with the help of hundreds of short DNA staple strands. This technique allowed for the creation of larger DNA constructs, increasing the size from 100-300 nucleotide pairs to over 7000, making it accessible to a wider community. Moreover, DNA origami simplified the process as, in most cases, the DNA did not need to be purified, just ordered from a supplier.
These advances in DNA nanotechnology have opened up new possibilities for the creation of nanoscale devices, sensors, and materials that could revolutionize many areas of science and technology.
DNA-based computing and data storage
DNA, or deoxyribonucleic acid, has the remarkable ability to store vast amounts of information by encoding it in sequences of CGAT nucleotides. The remarkable complexity and diversity of genetic codes across different species highlight the enormous potential for information storage using CGAT-encoded DNA, which can be leveraged in computing. Through a process called hybridization, where DNA pairs bond with each other, DNA molecules can be utilized to process information. This involves taking single DNA strands as input and transforming them into subsequent strands of DNA as output.
By synthesizing DNA molecules – making them from scratch – researchers have found they can specify, or write, long strings of the letters A, C, G, and T and then read those sequences back. The process is analogous to how a computer stores information using 0s and 1s. For example, you could use: 00 is A, 01 is C, 10 is G and 11 is T, and then based on that encode the entire sequence of zeros and ones in the code (read more: Translation software enables efficient DNA data storage).
DNA’s storage density is staggering. DNA molecules can store information many millions of times denser than existing technologies for digital storage – flash drives, hard drives, magnetic and optical media. Using DNA to directly store data is an attractive possibility because it is extremely dense, with a theoretical limit of 228 exabytes/mm3 of synthetic DNA, and long-lasting, with observed half-life of over 500 years. For instance, researchers already demonstrated they could store a 100 MB file of a TV show on a picogram – or a billionth of a gram of DNA. It would be theoretically possible to even store up to 228 exabytes on one gram of synthetic DNA.
Consider this: humanity will generate an estimated 33 zettabytes by 2025 – that’s 33 followed by 21 zeroes. If if becomes possible to utilize the full theoretical storage capacity of DNA, that information would require no more than 145 grams of DNA (we'll show you the detailed calculation for this below). Or in volume terms, this DNA archive would require avolume of roughly 8.5 x 10-5 cubic meters, or about 3 ping pong balls.
Here are the calculations if you want to delve deeper into this example:
How much information can be stored in DNA?
Human DNA is composed of approximately 3 billion base pairs. Using the 4 possible combinations of base pairs as a binary system, we can represent 2 bits of information with each base pair. To calculate the amount of information that can be stored in human DNA, we multiply the number of base pairs by the number of bits per base pair:
3,000,000,000 base pairs × 2 bits/base pair = 6,000,000,000 bits
To convert this value to bytes, we divide by 8:
6,000,000,000 bits / 8 bits/byte = 750,000,000 bytes
Thus, human DNA has a theoretical raw storage capacity of about 750 megabytes of information (not assuming the theoretical use of data compression techniques).
How much information can be stored in 1 gram of human DNA?
One human DNA molecule is composed of approximately 3 billion base pairs, and each base pair has an average molecular weight of 650 Daltons (Da).
Calculate the molecular weight of the entire human genome: 3,000,000,000 base pairs × 650 Da/base pair ≈ 1.95 × 10^12 Da
Convert Daltons to grams per mole using Avogadro's number (6.022 × 10^23): (1.95 × 10^12 Da) / (6.022 × 10^23 Da/mol) ≈ 3.24 × 10^-12 g/mol
Calculate the number of genomes in 1 gram: 1 g / (3.24 × 10^-12 g/mol) ≈ 3.08 × 10^11 genomes
Calculate the data storage capacity of 1 gram of DNA: (3.08 × 10^11 genomes) × (750 MB/genome) ≈ 2.31 × 10^14 MB
Thus, 1 gram of human DNA could theoretically store about 231 terabytes (TB) of raw data.
How much information can be stored in one gram of arbitrarily long synthetic DNA?
Calculate the weight of a single base pair in grams: 660 g/mol / 6.022 × 10^23 base pairs/mol ≈ 1.096 × 10^-21 g/base pair
Calculate the number of base pairs in 1 gram of DNA: 1 g / (1.096 × 10^-21 g/base pair) ≈ 9.12 × 10^20 base pairs
Calculate the number of bits that can be stored in these base pairs: (9.12 × 10^20 base pairs) × (2 bits/base pair) = 1.824 × 10^21 bits
Convert bits to bytes: 1.824 × 10^21 bits / 8 bits/byte = 2.28 × 10^20 bytes
Convert bytes to exabytes: 2.28 × 10^20 bytes / 10^18 bytes/exabyte ≈ 228 exabytes per gram
Therefore, 1 gram of an arbitrarily long synthetic DNA could theoretically store about 228 exabytes of data.
How much would 1 cubic millimeter of DNA weigh?
To calculate the weight of 1 cubic millimeter of DNA, we'll use the density of DNA that we mentioned earlier.
DNA density: The density of DNA is approximately 1.7 g/cm^3, or 1.7 g/mL.
To convert this to g/mm^3, we divide by 1000: 1.7 g/mL / 1000 = 0.0017 g/mm^3
Calculate the mass of DNA in 1 cubic millimeter: 0.0017 g/mm^3 × 1 mm^3 = 0.0017 g
Thus, 1 cubic millimeter of DNA would weigh approximately 0.0017 grams or 1.7 milligrams.
Information stored in DNA can be copied in a massively parallel manner and selectively retrieved via polymerase chain reaction (PCR). However, existing DNA storage systems suffer from high latency caused by the inherently sequential nature of the writing process. Despite recent progress, a typical cycle time of solid-phase DNA synthesis is on the order of minutes, which limits the practical applications of this molecular storage platform. To overcome these challenges, new synthesis methods and information encoding approaches are required to accelerate the speed of writing large-volume data sets (read more: An expanded molecular alphabet for DNA data storage).
DNA computing was first demonstrated in 1994 by Leonard Adleman who encoded and solved the travelling salesman problem, a mathematics problem to find the most efficient route for a salesman to take between hypothetical cities, entirely in DNA. This first prototype of a DNA computer was a test tube filled with 100 microliters of a DNA solution.
Ever since Adleman's experiment, various DNA-based 'circuits' have been proposed to execute computational methods such as Boolean logic, arithmetical formulas, and neural network computation. For instance, researchers have designed fuzzy and Boolean logic gates based on DNA: they synthesized self-assembled DNA complexes that sense two environmental signals and produce a fluorescent output corresponding to the operation of all six Boolean logic gates AND, NAND, OR, NOR, XOR, and XNOR.
This approach is called molecular programming, which utilizes computing concepts and designs in DNA nanotechnology, suitable for working with DNA at a nanoscale level.
Molecular programming is essentially biochemistry, where the ‘programs’ created are methods of selecting molecules that interact to achieve a specific result through DNA self-assembly. This process involves disordered collections of molecules spontaneously interacting to form the desired arrangement of DNA strands.
This development of DNA computing led to a significant impact on the DNA nanotechnology community, as computer scientists and mathematicians became aware of it and became major contributors to the field. This community shifted away from the crystallographer-based approach of building unit cells with tiles with sticky ends, and instead treated tiles as logic units.
DNA nanofabrication
DNA nanofabrication is a method for creating complex, precise nanostructures using DNA as a building material. It involves designing and synthesizing DNA molecules with specific sequences that can self-assemble into desired shapes and patterns.
At present, an ever-increasing number of research groups are exploiting programmable self-assembly properties of nucleic acids in creating rationally designed nanoshapes and nanomachines for many different uses.
These objects are similar in size to virus capsids and have gained a lot of interest as nanocontainers for drug delivery. Since they can be programed to carry out specific tasks (like transporting molecules and releasing them at a target site), these assemblies act like a robotic system – nanorobots (read more: The ongoing development of DNA nanotechnology).
Not to be confused with the nanorobots of science fiction stories, for medical nanotechnology researchers a nanorobot, or nanobot, is a popular term for molecules with a unique property that enables them to be programmed to carry out a specific task.
Designing synthetic DNA nanostructures
To design a DNA structure, scientists start with a desired shape or pattern and then determine the complementary DNA sequences that will allow the DNA strands to self-assemble into that structure. This involves carefully selecting the length and sequence of each DNA strand to ensure that it will bind to the appropriate complementary strand in the correct position.
The DNA strands – typically short synthetic DNA strands with 20-100 nucleotides in length – are synthesized using solid-phase DNA synthesis techniques. This involves adding nucleotides one at a time, building up the DNA strand in a controlled manner.
Once the DNA strands are synthesized and mixed together, they spontaneously self-assemble into the desired structure through complementary base pairing. This process is driven by the energetically favorable formation of hydrogen bonds between the complementary base pairs.
The resulting DNA structure is held together by a combination of hydrogen bonds, base stacking interactions, and electrostatic forces. The stability of the structure can be increased by introducing additional cross-links, such as covalent bonds or metal ions, between the DNA strands.
One powerful technique for creating complex DNA nanostructures is DNA origami. This involves using a long, single-stranded DNA molecule, known as a scaffold, to guide the folding of shorter DNA strands into a desired shape. The scaffold is designed with a specific sequence of base pairs that create a series of staple sites, where the shorter strands can bind to the scaffold and fold into the desired shape.
DNA origami
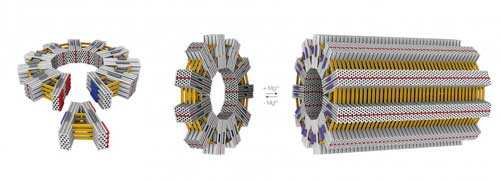
One of the most important developments in structural DNA nanotechnology has been the use of a scaffold strand and hundreds of short staple strands for the assembly of three-dimensional DNA origami objects.
For instance, by transferring viral construction principles to DNA origami technology, researchers can design and build structures on the scale of viruses and cell organelles (read more about this work here: DNA-origami surpasses important thresholds).
Applications of DNA nanotechnology
There are many exciting applications of DNA nanotechnology, some of which are already being developed and used in research. Here are a few examples:
Drug Delivery
DNA nanotechnology can be used to create drug delivery vehicles that are capable of targeting specific cells or tissues in the body. By designing and programming DNA structures with specific functional components, such as aptamers or antibodies, scientists can create nanoscale devices that can interact with biological systems in precise and controlled ways. See for instance: DNA folds into a smart nanocapsule for drug delivery.
Biosensors
DNA nanotechnology can be used to create biosensors that are capable of detecting specific molecules or pathogens in biological samples. By designing and programming DNA structures with specific recognition elements, such as aptamers or antibodies, scientists can create nanoscale devices that can bind to and detect specific molecules or pathogens. Such DNA biosensors could unlock powerful, low-cost clinical diagnostics.
Nanorobots
DNA nanotechnology can be used to create nanorobots that are capable of performing specific tasks in the body, such as delivering drugs or performing surgeries. By designing and programming DNA structures with specific functional components, such as motors or sensors, scientists can create nanoscale devices that can move and interact with biological systems in precise and controlled ways. Take for instance this example of a DNA origami nanorobot with a switchable flap.
DNA nanoarchitectonics
DNA nanotechnology can be used to create new materials with unique properties, such as high strength, elasticity, or conductivity. By designing and programming DNA structures with specific geometric and functional properties, scientists can create new materials with tailored properties for specific applications, such as energy storage or electronic devices. In an area termed DNA nanoarchitectonics, researchers are using assembled DNA as a nanoscale building block for interfacial layers. Applications range from thin-films and surface coatings, to bottom-up nanopatterning and 3D nanoparticle lattices.
Synthetic Biology
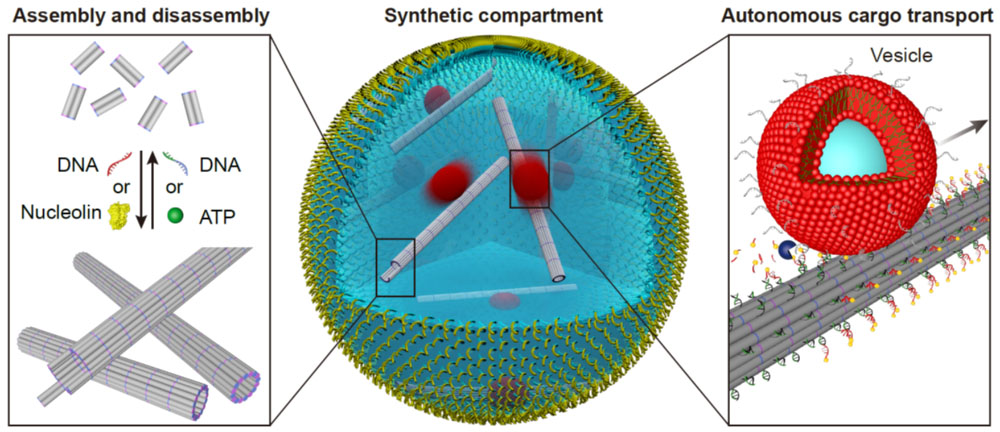
DNA nanotechnology can be used to create synthetic biological systems, such as synthetic cells or organisms. By designing and programming DNA structures with specific functional components, scientists can create nanoscale devices that can mimic the behavior of biological systems or create new biological functions.
For instance, building functional synthetic cells from the bottom-up is an ongoing effort of scientists around the globe. Already, researchers have taken steps to build artificial biological cells by using DNA-based filaments as a synthetic cytoskeleton and giving them diverse functionality.
Anti-counterfeiting with DNA nanotechnology
DNA-based protection technologies are especially suitable for anti-counterfeiting measures. For instance, DNA barcoding is a method of identifying and authenticating products using DNA sequences. In this technique, a unique DNA sequence is synthesized and attached to or included in a product, and the sequence is then recorded in a database. The DNA sequence can be easily read and compared to the database to verify the authenticity of the product. See an example here.
Frequently Asked Questions (FAQs) about DNA nanotechnology
What is DNA Nanotechnology?
DNA nanotechnology is an innovative field of study that utilizes the unique molecular recognition properties of DNA to create self-assembling nanostructures. These structures have a wide range of applications, from drug delivery systems to the construction of nanoscale machines.
How does DNA Nanotechnology work?
DNA nanotechnology works by taking advantage of the natural base pairing rules of DNA—adenine pairs with thymine and cytosine pairs with guanine—to design and synthesize specific sequences of DNA that will fold and self-assemble into predetermined shapes and structures.
What are the main applications of DNA Nanotechnology?
The applications of DNA nanotechnology are vast and varied, including drug delivery, biosensing, nanoelectronic circuits, data storage, and the creation of nanoscale robots. These applications leverage the programmability and predictability of DNA interactions.
How is DNA Nanotechnology different from traditional nanotechnology?
Unlike traditional nanotechnology, which often relies on top-down fabrication techniques, DNA nanotechnology employs a bottom-up approach. It uses the self-assembly properties of DNA to build structures from the molecular level up, allowing for more precise and programmable constructions.
What are the benefits of using DNA in nanotechnology?
DNA offers several benefits in nanotechnology, including biocompatibility, programmability, predictability, and the ability to self-assemble. These properties make DNA an ideal material for constructing complex nanostructures with high specificity and functionality.
Are there any ethical concerns associated with DNA Nanotechnology?
As with any emerging technology, DNA nanotechnology raises ethical questions, particularly concerning privacy, security, environmental impact, and the potential for misuse. Ongoing dialogue among scientists, ethicists, and policymakers is crucial to address these concerns.
How can DNA Nanotechnology impact healthcare?
DNA nanotechnology has the potential to revolutionize healthcare by enabling the development of highly targeted drug delivery systems, improved diagnostic tools, and novel therapies. This could lead to more effective treatments with fewer side effects.
What are the challenges facing DNA Nanotechnology?
Challenges include technical hurdles related to the stability and scalability of DNA nanostructures, as well as the need for further research to fully understand the long-term implications and behavior of DNA-based materials in biological systems.
How environmentally friendly is DNA Nanotechnology?
DNA nanotechnology is considered relatively environmentally friendly due to the biodegradability and biocompatibility of DNA. However, the environmental impact of large-scale production and application of DNA nanostructures needs further study.
What is the future of DNA Nanotechnology?
The future of DNA nanotechnology looks promising, with ongoing research pushing the boundaries of what is possible. Advances in computational design, precision manufacturing, and integration with other nanomaterials are expected to drive significant progress in the field.
Also check out our article on RNA nanotechnology.