Abstract
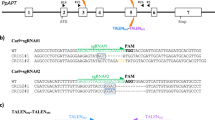
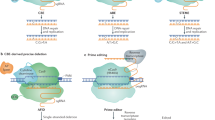
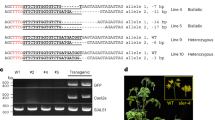
Gene expression is regulated by multiple processes, and the translation of mRNAs into proteins is an especially critical step. Upstream open reading frames (uORFs) are widespread cis-elements in eukaryotic genes that usually suppress the translation of downstream primary ORFs (pORFs). Here, we describe a protocol for fine-tuning gene translation in plants by editing endogenous uORFs with the CRISPR–Cas9 system. The method we present readily yields transgene-free uorf mutant offspring. We provide detailed protocols for predicting uORFs and testing their effects on downstream pORFs using a dual-luciferase reporter system, designing and constructing single guide RNA (sgRNA)–Cas9 vectors, identifying transgene-free uorf mutants, and finally comparing the mRNA, protein and phenotypic levels of target genes in uorf mutants and controls. Predicting uORFs and confirming their effects in protoplasts takes only 2–3 weeks, and transgene-free mutants with edited target uORFs controlling different levels of pORF translation can be obtained within 4 months. Unlike previous methods, our strategy achieves fine-tuning of gene translation in transgene-free derivatives, which accelerates the analysis of gene function and the improvement of crop traits.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The authors declare that all data supporting the findings of this study are available in the original paper. Other supporting data are available upon reasonable request to the corresponding author.
References
de Montaigu, A. et al. Natural diversity in daily rhythms of gene expression contributes to phenotypic variation. Proc. Natl Acad. Sci. USA 112, 905–910 (2015).
Butaye, K. M. J., Cammue, B. P. A., Delaure, S. L. & De Bolle, M. F. C. Approaches to minimize variation of transgene expression in plants. Mol. Breed. 16, 79–91 (2005).
Hayama, R., Yokoi, S., Tamaki, S., Yano, M. & Shimamoto, K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722 (2003).
Ito, T. & Meyerowitz, E. M. Overexpression of a gene encoding a cytochrome p450,CYP78A9, induces large and seedless fruit in Arabidopsis. Plant Cell 12, 1541–1550 (2000).
Lowder, L. G. et al. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169, 971–985 (2015).
Rodriguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E. & Lippman, Z. B. Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480 (2017).
Schwanhausser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Hellens, R. P., Brown, C. M., Chisnall, M. A. W., Waterhouse, P. M. & Macknight, R. C. The emerging world of small ORFs. Trends Plant Sci. 21, 317–328 (2016).
Zhang, H., Wang, Y. & Lu, J. Function and evolution of upstream ORFs in eukaryotes. Trends Biochem. Sci. 44, 782–794 (2019).
Wethmar, K. The regulatory potential of upstream open reading frames in eukaryotic gene expression. Wiley Interdiscip. Rev. RNA 5, 765–778 (2014).
von Arnim, A. G., Jia, Q. & Vaughn, J. N. Regulation of plant translation by upstream open reading frames. Plant Sci. 214, 1–12 (2014).
Chew, G. L., Pauli, A. & Schier, A. F. Conservation of uORF repressiveness and sequence features in mouse, human and zebrafish. Nat. Commun. 7, 11663 (2016).
Liang, X. H. et al. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat. Biotechnol. 34, 875–880 (2016).
Xu, G. et al. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 545, 491–494 (2017).
Knott, G. J. & Doudna, J. A. CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869 (2018).
Chen, K., Wang, Y., Zhang, R., Zhang, H. & Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697 (2019).
Pawelczak, K. S., Gavande, N. S., VanderVere-Carozza, P. S. & Turchi, J. J. Modulating DNA repair pathways to improve precision genome engineering. ACS Chem. Biol. 13, 389–396 (2017).
Ran, Y., Liang, Z. & Gao, C. Current and future editing reagent delivery systems for plant genome editing. Sci. China Life Sci. 60, 490–505 (2017).
Zhang, Y. et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617 (2016).
Liang, Z. et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8, 14261 (2017).
Woo, J. W. et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164 (2015).
Osakabe, Y. et al. CRISPR-Cas9-mediated genome editing in apple and grapevine. Nat. Protoc. 13, 2844–2863 (2018).
Zhang, H. et al. Genome editing of upstream open reading frames enables translational control in plants. Nat. Biotechnol. 36, 894–898 (2018).
Li, T. et al. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36, 1160–1163 (2018).
Zong, Y. et al. Precise base editing in rice, wheat and maize with a Cas9-cytidinedeaminase fusion. Nat. Biotechnol. 35, 438–440 (2017).
Zong, Y. et al. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 36, 950–953 (2018).
Chen, Y. et al. CRISPR/Cas9-mediated base-editing system efficiently generatesgain-of-function mutations in Arabidopsis. Sci. China Life Sci. 60, 520–523 (2017).
Xue, C., Zhang, H., Lin, Q., Fan, R. & Gao, C. Manipulating mRNA splicing by base editing in plants. Sci. China Life Sci. 61, 1293–1300 (2018).
Li, C. et al. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19, 59 (2018).
Tak, Y. E. et al. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat. Methods 14, 1163–1166 (2017).
Sidorenko, L. V. et al. GC-rich coding sequences reduce transposon-like, small RNA-mediated transgene silencing. Nat. Plants 3, 875–884 (2017).
Jones, H. D. Regulatory uncertainty over genome editing. Nat. Plants 1, 14011 (2015).
Li, Z. et al. A potent Cas9-derived gene activator for plant and mammalian cells. Nat. Plants 3, 930–936 (2017).
Sarkar, A. K. et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814 (2007).
Khanday, I., Skinner, D., Yang, B., Mercier, R. & Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565, 91–95 (2019).
Pi, L. et al. Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell 33, 576–588 (2015).
Hinnebusch, A. G., Ivanov, I. P. & Sonenberg, N. Translational control by 5’-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416 (2016).
Laing, W. A. et al. An upstream open reading frame is essential for feedback regulation of ascorbate biosynthesis in Arabidopsis. Plant Cell 27, 772–786 (2015).
Merchante, C., Stepanova, A. N. & Alonso, J. M. Translation regulation in plants: an interesting past, an exciting present and a promising future. Plant J. 90, 628–653 (2017).
Jorgensen, R. A. & Dorantes-Acosta, A. E. Conserved peptide upstream open reading frames are associated with regulatory genes in angiosperms. Front. Plant Sci. 3, 191 (2012).
Hellens, R. P. et al. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13 (2005).
Yan, L. et al. High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8, 1820–1823 (2015).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR Cas system. Cell 163, 759–771 (2015).
Endo, M. et al. Genome editing in plants by engineered CRISPR-Cas9 recognizing NG PAM. Nat. Plants 5, 14–17 (2019).
Hu, J. H. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018).
Zhang, X., Henriques, R., Lin, S. S., Niu, Q. W. & Chua, N. H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646 (2006).
Shan, Q., Wang, Y., Li, J. & Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 9, 2395–2410 (2014).
Liang, Z. et al. Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 13, 413–430 (2018).
Bae, S., Park, J. & Kim, J. S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Xing, H. L. et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327 (2014).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Zhai, Z., Jung, H. I. & Vatamaniuk, O. K. Isolation of protoplasts from tissues of 14-day-old seedlings of Arabidopsis thaliana. J. Vis. Exp. 30, e1149 (2009).
Xu, C. et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47, 784–792 (2015).
Nolan, T., Hands, R. E. & Bustin, S. A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1, 1559–1582 (2006).
Wang, Z. Y., Seto, H., Fujioka, S., Yoshida, S. & Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383 (2001).
Espinosa-Ruiz, A., Martínez, C. & Prat, S. Protocol to treat seedlings with brassinazole and measure hypocotyl length in Arabidopsis thaliana. Bio Protoc. 5, e1568 (2015).
Kovács, L. et al. Quantitative determination of ascorbate from the green alga Chlamydomonas reinhardtii by HPLC. Bio Protoc. 6, e2067 (2016).
Lowder, L., Malzahn, A. & Qi, Y. Rapid evolution of manifold CRISPR systems for plant genome editing. Front. Plant Sci. 7, 1683 (2016).
Höfgen, R. & Willmitzer, L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16, 9877 (1988).
Acknowledgements
This work was supported by grants from the National Transgenic Science and Technology Program (2018ZX0801002B, 2019ZX08010-001, 2018ZX0800102B and 2019ZX08010-003) and the National Natural Science Foundation of China (31788103 and 31971370).
Author information
Authors and Affiliations
Contributions
X.S. performed the experiments; Y.W., X.S. and K.C. designed figures; C.G. supervised the project; X.S., H.Z. and C.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Jian Lu, Yiping Qi, Changfu Zhu, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Zhang, H. et al. Nat. Biotechnol. 36, 894–898 (2018): https://doi.org/10.1038/nbt.4202
Li, T. et al. Nat. Biotechnol. 36, 1160–1163 (2018): https://doi.org/10.1038/nbt.4273
Supplementary information
Supplementary Information
Supplementary Notes 1 and 2 and Supplementary Table 1.
Rights and permissions
About this article
Cite this article
Si, X., Zhang, H., Wang, Y. et al. Manipulating gene translation in plants by CRISPR–Cas9-mediated genome editing of upstream open reading frames. Nat Protoc 15, 338–363 (2020). https://doi.org/10.1038/s41596-019-0238-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0238-3
This article is cited by
-
Application of genome editing techniques to regulate gene expression in crops
BMC Plant Biology (2024)
-
OsMADS17 simultaneously increases grain number and grain weight in rice
Nature Communications (2023)
-
Tuning plant phenotypes by precise, graded downregulation of gene expression
Nature Biotechnology (2023)
-
Trait Improvement of Solanaceae Fruit Crops for Vertical Farming by Genome Editing
Journal of Plant Biology (2023)
-
Fine-tuning of quantitative traits
Science China Life Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.