ADME Gene Regulation by ncRNAs
ncRNAs, particularly miRNAs, have been considered to play a significant role in ADME gene regulation. Many miRNAs have also been predicted to target numerous ADME-encoding genes in silico.[41] However, the list of ADME genes that have been experimentally validated by typical reporter gene assays as targets of specific miRNAs is much shorter. These ADME genes, the respective miRNAs involved in their regulation and the cellular systems used in validation studies are summarized in Table 2. In most cases, miRNAs typically directly target the 3'-UTR region of the ADME gene in question; however, ADME genes can also be significantly affected by indirect miRNA targeting of the transcription factors that tightly regulate their expression. For example, the direct regulation of PXR by miR148 and VDR by miR27B can affect the expression of CYP3A4. Moreover, the regulation of ADME genes by miRNA has implications in the pharmacology of drugs. For a more comprehensive review on miRNA regulation of ADME genes and some related pharmacological consequences we refer readers to recent reviews in this field.[42–46]
In addition to miRNAs, the potential for recently described lncRNAs in the regulation of ADME genes is highly plausible. For example, evidence in mice suggests that the lncRNA Air can negatively regulate the expression of genes encoding the drug transporters Slc22a3 and Slc22a3 through targeted recruitment of repressive histone-modifications.[47]Air is a paternal-specific ncRNA on mouse chromosome 17 that is expressed from a promoter located in intron 2 of the Igf2r gene. This 108-kb ncRNA has a 3.7-kb imprint control element containing its promoter. It releases paternal-specific silencing of the Igf2r gene and the Slc22a2 and Slc22a3 genes, which are located upstream and not overlapping or sharing DNA sequence homology with Air.[48] Silencing by Air is also regulated in a tissue- and developmental-specific manner in the brain and placenta, the basis of which remains unknown.[49,50] Although evidence of ADME gene regulation is not yet available in humans, these emerging findings demonstrate the additional epigenetic layer of complex gene regulation mediated by lncRNAs that is plausible to extend to the transcriptional control of ADME gene expression. Possible and confirmed mechanisms of epigenetic control of ADME gene expression are illustrated in Figure 1.
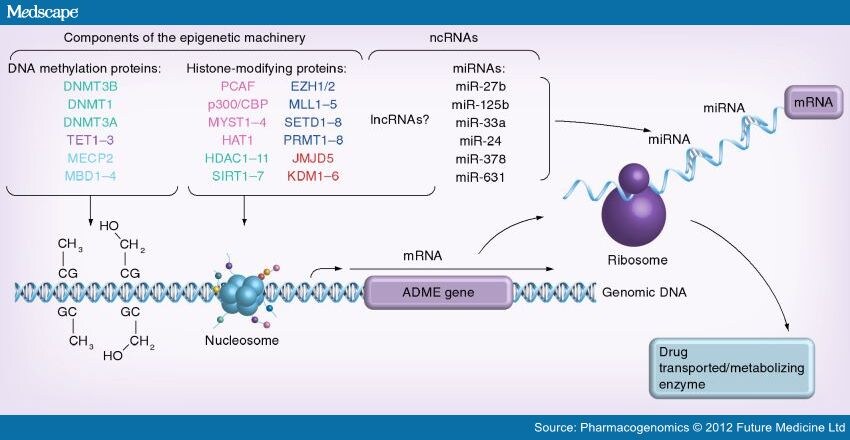
Figure 1.
Possible mechanisms of epigenetic control of absorption, distribution, metabolism and excretion gene expression.
Multiple epigenetic enzymes are responsible for DNA methylation/hydroxymethylation and various histone modifications. DNA methylation-modifying enzymes include: DNA methyltransferases DNMT1, DNMT3A and DNMT3B (converting cytosine residues into 5-methylcytosine) and dioxygenases TET1–3, responsible for the hydroxymethylation of 5mC. Histone-modifying enzymes include histone lysine acetyltransferases (p300/CBP, NCOA1/3, MYST1–4 and PCAF) and deacetylases (HDAC1–11 and SIRT1–7); histone lysine (EZH1/2, MLL1–5 and SETD1–8, among others) and arginine (PRMT1–8) methyltransferases, as well as lysine (KDM1–6 and JMJD5, among others) and arginine demethylases (JMJD6); multiple histone serine/threonine/tyrosine kinases and phosphatases, as well as histone lysine biotinases, (de)ribosylases, (de)ubiquitinases and arginine deiminases. MBD1–4 and MECP2 are specific proteins which recognize and bind to methylated DNA or nucleosomes manifesting a specific combination of histone-modification patterns ('the histone code'). Taken together, these factors define the chromatin confirmation in a given genomic locus as transcriptionally active ('open') or inactive ('closed') epigenetic state. In addition, lncRNAs are presumed to recognize specific genomic loci and recruit histone-modifying enzymes to the targeted genes thus establishing epigenetic marks in gene promoters. This orchestrated network of epigenetic enzymes along with ncRNAs can potentially affect the expression of ADME genes. Epigenetic changes in ADME genes can in turn contribute to interindividual differences in mRNA levels of these genes. Furthermore, miRNAs can additionally participate in the post-transcriptional ADME gene regulation and contribute to expression levels of ADME enzymes. Hence, interindividual differences in the activity of ADME enzymes can originate from specific epigenetic marks located in these genes as well from certain ncRNA-dependent regulation.
ADME: Absorption, distribution, metabolism and excretion.
Pharmacogenomics. 2012;13(12):1373-1385. © 2012 Future Medicine Ltd.