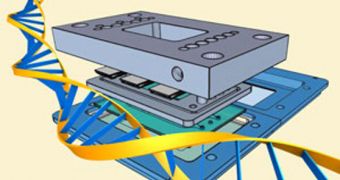
DNA sequencing is a very important tool in medicine and biomedical research, as it plays a very important role in investigations related to studying genetic diseases and also cancer. But existing technologies aimed at sequencing can be very complex, and require a lot of time and machines to be conducted. This makes their costs jump through the roof, and therefore unfordable to most. But, now, American researchers have compiled a new sequencing method, which requires no more than a DNA sample, and a chip featuring an integrated microfluidic device, Chemistry World reports.
The method, called sequencing by denaturation (SBD), has been developed by researchers at the University of California in San Diego (UCSD), led by expert Xiaohua Huang. Rather than sequencing only one or two DNA bases at a time, and repeating the cycle, their technique revolves around randomly injecting fluorescent nucleotides into the sample of genetic material. As this happens, the double-helix strand is broken apart in pieces of various lengths, each of them with its own fluorescent nucleotid, tied to its corresponding base type. Depending on length, the strands that are formed in this process have various melting temperatures.
After the first step is completed, the strands are heated in a solution, which causes them to start denaturating, one by one. The shorter ones go first, while the longer ones take a bit more time before they disappear as well. The researchers keep a close eye on the way the fluorescence of the solution decreases during this heating stage. The signal emitted by the DNA strands as they go can be analyzed to determine the base sequence of the target DNA template. This enables the scientists to make sequencing faster, cheaper and less demanding than it has ever been.
“This device has the capability for performing high-speed fluorescence imaging while biomedical reactions with precisely controlled temperature profiles take place. Although the read length is quite limited, due to its simplicity, SBD has the potential to bring down the cost of large-scale sequencing drastically,” Huang adds. “I would think that this approach would be very useful for genotyping in the future where full sequencing is not necessary,” University of New Mexico in Alberquerque functional genomics tool development expert Jeremy Edwards says.
 14 DAY TRIAL //
14 DAY TRIAL //