Abstract
Genetically engineered T cell immunotherapies have provided remarkable clinical success to treat B cell acute lymphoblastic leukaemia by harnessing a patient’s own T cells to kill cancer, and these approaches have the potential to provide therapeutic benefit for numerous other cancers, infectious diseases and autoimmunity. By introduction of either a transgenic T cell receptor or a chimeric antigen receptor, T cells can be programmed to target cancer cells. However, initial studies have made it clear that the field will need to implement more complex levels of genetic regulation of engineered T cells to ensure both safety and efficacy. Here, we review the principles by which our knowledge of genetics and genome engineering will drive the next generation of adoptive T cell therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Rappuoli, R., Mandl, C. W., Black, S. & De Gregorio, E. Vaccines for the twenty-first century society. Nat. Rev. Immunol. 11, 865–872 (2011).
Sliwkowski, M. X. & Mellman, I. Antibody therapeutics in cancer. Science 341, 1192–1198 (2013).
Pardoll, D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012).
Feldmann, M. & Maini, R. N. Anti-TNF alpha therapy of rheumatoid arthritis: what have we learned? Annu. Rev. Immunol. 19, 163–196 (2001).
Kumar, B. V., Connors, T. J. & Farber, D. L. Human T cell development, localization, and function throughout life. Immunity 48, 202–213 (2018).
Jameson, S. C. & Masopust, D. Understanding subset diversity in T cell memory. Immunity 48, 214–226 (2018).
Francica, J. R. et al. Steric shielding of surface epitopes and impaired immune recognition induced by the ebola virus glycoprotein. PLoS Pathog. 6, e1001098 (2010).
Wei, F. et al. Strength of PD-1 signaling differentially affects T-cell effector functions. Proc. Natl Acad. Sci. USA 110, E2480–E2489 (2013).
Park, J. H., Geyer, M. B. & Brentjens, R. J. CD19-targeted CAR T-cell therapeutics for hematologic malignancies: interpreting clinical outcomes to date. Blood 127, 3312–3320 (2016).
Stadtmauer, E. A. et al. CRISPR-engineered T cells in patients with refractory cancer. Science 367, eaba7365 (2020). This is the first report on the in vivo fate and genomic ramifications of CRISPR-engineered TCR T cells in a phase I clinical trial.
Li, J., Hong, S., Chen, W., Zuo, E. & Yang, H. Advances in detecting and reducing off-target effects generated by CRISPR-mediated genome editing. J. Genet. Genomics 46, 513–521 (2019).
Maldini, C. R., Ellis, G. I. & Riley, J. L. CAR T cells for infection, autoimmunity and allotransplantation. Nat. Rev. Immunol. 18, 605–616 (2018).
Rosado-Sanchez, I. & Levings, M. K. Building a CAR-Treg: going from the basic to the luxury model. Cell Immunol. 358, 104220 (2020).
Klichinsky, M. et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 38, 947–953 (2020).
Kimbrel, E. A. & Lanza, R. Next-generation stem cells - ushering in a new era of cell-based therapies. Nat. Rev. Drug Discov. 19, 463–479 (2020).
Hartweger, H. et al. HIV-specific humoral immune responses by CRISPR/Cas9-edited B cells. J. Exp. Med. 216, 1301–1310 (2019).
Fesnak, A. D., June, C. H. & Levine, B. L. Engineered T cells: the promise and challenges of cancer immunotherapy. Nat. Rev. Cancer 16, 566–581 (2016).
Newrzela, S. et al. T-cell receptor diversity prevents T-cell lymphoma development. Leukemia 26, 2499–2507 (2012).
Scholler, J. et al. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci. Transl Med. 4, 132ra153 (2012).
O’Leary, M. C. et al. FDA approval summary: tisagenlecleucel for treatment of patients with relapsed or refractory B-cell precursor acute lymphoblastic leukemia. Clin. Cancer Res. 25, 1142–1146 (2019).
Bouchkouj, N. et al. FDA approval summary: axicabtagene ciloleucel for relapsed or refractory large B-cell lymphoma. Clin. Cancer Res. 25, 1702–1708 (2019).
Hirayama, A. V. et al. High rate of durable complete remission in follicular lymphoma after CD19 CAR-T cell immunotherapy. Blood 134, 636–640 (2019).
Schuster, S. J. et al. Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N. Engl. J. Med. 380, 45–56 (2019).
Maude, S. L. et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018).
Locke, F. L. et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1-2 trial. Lancet Oncol. 20, 31–42 (2019).
Fiorenza, S., Ritchie, D. S., Ramsey, S. D., Turtle, C. J. & Roth, J. A. Value and affordability of CAR T-cell therapy in the United States. Bone Marrow Transplant. 55, 1706–1715 (2020).
Kagoya, Y. et al. Genetic ablation of HLA class I, class II, and the T-cell receptor enables allogeneic T cells to be used for adoptive T-cell therapy. Cancer Immunol. Res. 8, 926–936 (2020).
Cruz, C. R. et al. Infusion of donor-derived CD19-redirected virus-specific T cells for B-cell malignancies relapsed after allogeneic stem cell transplant: a phase 1 study. Blood 122, 2965–2973 (2013).
Kochenderfer, J. N. et al. Donor-derived CD19-targeted T cells cause regression of malignancy persisting after allogeneic hematopoietic stem cell transplantation. Blood 122, 4129–4139 (2013).
Huls, M. H. et al. Clinical application of Sleeping Beauty and artificial antigen presenting cells to genetically modify T cells from peripheral and umbilical cord blood. J. Vis. Exp. https://doi.org/10.3791/50070 (2013).
Pegram, H. J. et al. IL-12-secreting CD19-targeted cord blood-derived T cells for the immunotherapy of B-cell acute lymphoblastic leukemia. Leukemia 29, 415–422 (2015).
Ueda, T. & Kaneko, S. In vitro differentiation of T cell: from CAR-modified T-iPSC. Methods Mol. Biol. 2048, 85–91 (2019).
Themeli, M. et al. Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nat. Biotechnol. 31, 928–933 (2013).
Yan, L. Z. et al. Allogeneic CAR-T for treatment of relapsed and/or refractory multiple myeloma: four cases report and literatures review. Zhonghua Xue Ye Xue Za Zhi 40, 650–655 (2019).
Brudno, J. N. et al. Allogeneic T cells that express an anti-CD19 chimeric antigen receptor induce remissions of B-cell malignancies that progress after allogeneic hematopoietic stem-cell transplantation without causing graft-versus-host disease. J. Clin. Oncol. 34, 1112–1121 (2016).
Eapen, M. et al. Effect of graft source on unrelated donor haemopoietic stem-cell transplantation in adults with acute leukaemia: a retrospective analysis. Lancet Oncol. 11, 653–660 (2010).
Kwoczek, J. et al. Cord blood-derived T cells allow the generation of a more naive tumor-reactive cytotoxic T-cell phenotype. Transfusion 58, 88–99 (2018).
Torikai, H. et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood 119, 5697–5705 (2012).
Stenger, D. et al. Endogenous TCR promotes in vivo persistence of CD19-CAR-T cells compared to a CRISPR/Cas9-mediated TCR knockout CAR. Blood 136, 1407–1418 (2020).
Steentoft, C. et al. Glycan-directed CAR-T cells. Glycobiology 28, 656–669 (2018).
Hughes, M. S. et al. Transfer of a TCR gene derived from a patient with a marked antitumor response conveys highly active T-cell effector functions. Hum. Gene Ther. 16, 457–472 (2005).
Zhao, Y. et al. Primary human lymphocytes transduced with NY-ESO-1 antigen-specific TCR genes recognize and kill diverse human tumor cell lines. J. Immunol. 174, 4415–4423 (2005).
Ahmadi, M. et al. CD3 limits the efficacy of TCR gene therapy in vivo. Blood 118, 3528–3537 (2011).
van Loenen, M. M. et al. Mixed T cell receptor dimers harbor potentially harmful neoreactivity. Proc. Natl Acad. Sci. USA 107, 10972–10977 (2010).
Shao, H. et al. TCR mispairing in genetically modified T cells was detected by fluorescence resonance energy transfer. Mol. Biol. Rep. 37, 3951–3956 (2010).
Cohen, C. J. et al. Enhanced antitumor activity of T cells engineered to express T-cell receptors with a second disulfide bond. Cancer Res. 67, 3898–3903 (2007).
Bendle, G. M. et al. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat. Med. 16, 565–570 (2010).
Govers, C., Sebestyen, Z., Coccoris, M., Willemsen, R. A. & Debets, R. T cell receptor gene therapy: strategies for optimizing transgenic TCR pairing. Trends Mol. Med. 16, 77–87 (2010).
Yang, S. et al. Development of optimal bicistronic lentiviral vectors facilitates high-level TCR gene expression and robust tumor cell recognition. Gene Ther. 15, 1411–1423 (2008).
Legut, M., Dolton, G., Mian, A. A., Ottmann, O. G. & Sewell, A. K. CRISPR-mediated TCR replacement generates superior anticancer transgenic T cells. Blood 131, 311–322 (2018).
Okamoto, S. et al. Improved expression and reactivity of transduced tumor-specific TCRs in human lymphocytes by specific silencing of endogenous TCR. Cancer Res. 69, 9003–9011 (2009).
Robbins, P. F. et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol. 29, 917–924 (2011).
Maldini, C. R. et al. Dual CD4-based CAR T cells with distinct costimulatory domains mitigate HIV pathogenesis in vivo. Nat. Med. 26, 1776–1787 (2020).
Guedan, S. et al. ICOS-based chimeric antigen receptors program bipolar TH17/TH1 cells. Blood 124, 1070–1080 (2014).
Kawalekar, O. U. et al. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity 44, 380–390 (2016).
Liu, X. et al. Affinity-tuned ErbB2 or EGFR chimeric antigen receptor T cells exhibit an increased therapeutic index against tumors in mice. Cancer Res. 75, 3596–3607 (2015).
Ghorashian, S. et al. Enhanced CAR T cell expansion and prolonged persistence in pediatric patients with ALL treated with a low-affinity CD19 CAR. Nat. Med. 25, 1408–1414 (2019).
Ellebrecht, C. T. et al. Reengineering chimeric antigen receptor T cells for targeted therapy of autoimmune disease. Science 353, 179–184 (2016).
Brusko, T. M., Wasserfall, C. H., Clare-Salzler, M. J., Schatz, D. A. & Atkinson, M. A. Functional defects and the influence of age on the frequency of CD4+ CD25+ T-cells in type 1 diabetes. Diabetes 54, 1407–1414 (2005).
Thiruppathi, M. et al. Functional defect in regulatory T cells in myasthenia gravis. Ann. N. Y. Acad. Sci. 1274, 68–76 (2012).
Viglietta, V., Baecher-Allan, C., Weiner, H. L. & Hafler, D. A. Loss of functional suppression by CD4+CD25+ regulatory T cells in patients with multiple sclerosis. J. Exp. Med. 199, 971–979 (2004).
Brunstein, C. G. et al. Umbilical cord blood-derived T regulatory cells to prevent GVHD: kinetics, toxicity profile, and clinical effect. Blood 127, 1044–1051 (2016).
Plesa, G. et al. TCR affinity and specificity requirements for human regulatory T-cell function. Blood 119, 3420–3430 (2012).
Boardman, D. A. et al. Expression of a chimeric antigen receptor specific for donor HLA class I enhances the potency of human regulatory T cells in preventing human skin transplant rejection. Am. J. Transpl. 17, 931–943 (2017).
MacDonald, K. G. et al. Alloantigen-specific regulatory T cells generated with a chimeric antigen receptor. J. Clin. Invest. 126, 1413–1424 (2016). This study demonstrates that human CAR Treg cells can prevent GVHD in vivo.
Noyan, F. et al. Prevention of allograft rejection by use of regulatory T cells with an MHC-specific chimeric antigen receptor. Am. J. Transpl. 17, 917–930 (2017).
Dawson, N. A. et al. Systematic testing and specificity mapping of alloantigen-specific chimeric antigen receptors in regulatory T cells. JCI Insight 4, e123672 (2019).
Yeh, W. I. et al. Avidity and bystander suppressive capacity of human regulatory T cells expressing de novo autoreactive T-cell receptors in type 1 diabetes. Front. Immunol. 8, 1313 (2017).
Hull, C. M. et al. Generation of human islet-specific regulatory T cells by TCR gene transfer. J. Autoimmun. 79, 63–73 (2017).
Dawson, N. A. J. et al. Functional effects of chimeric antigen receptor co-receptor signaling domains in human regulatory T cells. Sci. Transl Med. 12, eaaz3866 (2020).
Boroughs, A. C. et al. Chimeric antigen receptor costimulation domains modulate human regulatory T cell function. JCI Insight 5, e126194 (2019).
Hippen, K. L. et al. Massive ex vivo expansion of human natural regulatory T cells (Tregs) with minimal loss of in vivo functional activity. Sci. Transl Med. 3, 83ra41 (2011).
McKenna, D. H., Jr. et al. Optimization of cGMP purification and expansion of umbilical cord blood-derived T-regulatory cells in support of first-in-human clinical trials. Cytotherapy 19, 250–262 (2017).
Zhou, X. et al. Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat. Immunol. 10, 1000–1007 (2009).
Honaker, Y. et al. Gene editing to induce FOXP3 expression in human CD4+ T cells leads to a stable regulatory phenotype and function. Sci. Transl Med. 12, eaay6422 (2020).
Kumar, M., Keller, B., Makalou, N. & Sutton, R. E. Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 12, 1893–1905 (2001).
Mitchell, R. S. et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2, E234 (2004).
Yang, O. O. et al. Lysis of HIV-1-infected cells and inhibition of viral replication by universal receptor T cells. Proc. Natl Acad. Sci. USA 94, 11478–11483 (1997).
Deeks, S. G. et al. A phase II randomized study of HIV-specific T-cell gene therapy in subjects with undetectable plasma viremia on combination antiretroviral therapy. Mol. Ther. 5, 788–797 (2002).
Mitsuyasu, R. T. et al. Prolonged survival and tissue trafficking following adoptive transfer of CD4zeta gene-modified autologous CD4+ and CD8+ T cells in human immunodeficiency virus-infected subjects. Blood 96, 785–793 (2000).
Leibman, R. S. et al. Supraphysiologic control over HIV-1 replication mediated by CD8 T cells expressing a re-engineered CD4-based chimeric antigen receptor. PLoS Pathog. 13, e1006613 (2017).
Bobisse, S. et al. Reprogramming T lymphocytes for melanoma adoptive immunotherapy by T-cell receptor gene transfer with lentiviral vectors. Cancer Res. 69, 9385–9394 (2009).
Zufferey, R., Donello, J. E., Trono, D. & Hope, T. J. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 73, 2886–2892 (1999).
Norrman, K. et al. Quantitative comparison of constitutive promoters in human ES cells. PLoS ONE 5, e12413 (2010).
Eyquem, J. et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543, 113–117 (2017). The authors improve CAR function by CRISPR-mediated insertion of CAR into the TRAC gene, improving expression kinetics and decreasing exhaustion.
Masiuk, K. E., Laborada, J., Roncarolo, M. G., Hollis, R. P. & Kohn, D. B. Lentiviral gene therapy in HSCs restores lineage-specific Foxp3 expression and suppresses autoimmunity in a mouse model of IPEX syndrome. Cell Stem Cell 24, 309–317.e7 (2019).
Fraietta, J. A. et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558, 307–312 (2018). Random insertion of CAR into the TET2 locus highlights the potential for targeting the epigenome to improve CAR T cell function and the value of analysis of the clinical product.
Zhao, Y. et al. High-efficiency transfection of primary human and mouse T lymphocytes using RNA electroporation. Mol. Ther. 13, 151–159 (2006).
Zhao, Y. et al. Multiple injections of electroporated autologous T cells expressing a chimeric antigen receptor mediate regression of human disseminated tumor. Cancer Res. 70, 9053–9061 (2010).
Jin, Z. et al. The hyperactive sleeping beauty transposase SB100X improves the genetic modification of T cells to express a chimeric antigen receptor. Gene Ther. 18, 849–856 (2011).
Chicaybam, L. et al. Transposon-mediated generation of CAR-T cells shows efficient anti B-cell leukemia response after ex vivo expansion. Gene Ther. 27, 85–95 (2020).
Gogol-Doring, A. et al. Genome-wide profiling reveals remarkable parallels between insertion site selection properties of the MLV retrovirus and the Piggybac transposon in primary human CD4+ T cells. Mol. Ther. 24, 592–606 (2016).
Berry, C., Hannenhalli, S., Leipzig, J. & Bushman, F. D. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput. Biol. 2, e157 (2006).
Chicaybam, L. et al. CAR T cells generated using Sleeping Beauty transposon vectors and expanded with an EBV-transformed lymphoblastoid cell line display antitumor activity in vitro and in vivo. Hum. Gene Ther. 30, 511–522 (2019).
Voigt, F. et al. Sleeping Beauty transposase structure allows rational design of hyperactive variants for genetic engineering. Nat. Commun. 7, 11126 (2016).
Querques, I. et al. A highly soluble Sleeping Beauty transposase improves control of gene insertion. Nat. Biotechnol. 37, 1502–1512 (2019).
Monjezi, R. et al. Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors. Leukemia 31, 186–194 (2017).
Kebriaei, P. et al. Phase I trials using sleeping beauty to generate CD19-specific CAR T cells. J. Clin. Invest. 126, 3363–3376 (2016).
Magnani, C. F. et al. Sleeping Beauty-engineered CAR T cells achieve antileukemic activity without severe toxicities. J. Clin. Invest. 130, 6021–6033 (2020).
Wang, J. et al. Highly efficient homology-driven genome editing in human T cells by combining zinc-finger nuclease mRNA and AAV6 donor delivery. Nucleic Acids Res. 44, e30 (2016).
MacLeod, D. T. et al. Integration of a CD19 CAR into the TCR alpha chain locus streamlines production of allogeneic gene-edited CAR T cells. Mol. Ther. 25, 949–961 (2017).
Hubbard, N. et al. Targeted gene editing restores regulated CD40L function in X-linked hyper-IgM syndrome. Blood 127, 2513–2522 (2016).
Sather, B. D. et al. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Transl Med. 7, 307ra156 (2015).
Roth, T. L. et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 559, 405–409 (2018).
Nguyen, D. N. et al. Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat. Biotechnol. 38, 44–49 (2020).
Sadelain, M., Papapetrou, E. P. & Bushman, F. D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer 12, 51–58 (2011).
Hale, M. et al. Engineering HIV-resistant, anti-HIV chimeric antigen receptor T cells. Mol. Ther. 25, 570–579 (2017).
Park, J. H. et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med. 378, 449–459 (2018).
Milone, M. C. et al. Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Mol. Ther. 17, 1453–1464 (2009).
Morgan, P. et al. Can the flow of medicines be improved? Fundamental pharmacokinetic and pharmacological principles toward improving phase II survival. Drug Discov. Today 17, 419–424 (2012).
Ager, A., Watson, H. A., Wehenkel, S. C. & Mohammed, R. N. Homing to solid cancers: a vascular checkpoint in adoptive cell therapy using CAR T-cells. Biochem. Soc. Trans. 44, 377–385 (2016).
Buckanovich, R. J. et al. Endothelin B receptor mediates the endothelial barrier to T cell homing to tumors and disables immune therapy. Nat. Med. 14, 28–36 (2008).
Motz, G. T. et al. Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat. Med. 20, 607–615 (2014).
Yamamoto, T. N. et al. T cells genetically engineered to overcome death signaling enhance adoptive cancer immunotherapy. J. Clin. Invest. 129, 1551–1565 (2019).
Lu, P., Weaver, V. M. & Werb, Z. The extracellular matrix: a dynamic niche in cancer progression. J. Cell Biol. 196, 395–406 (2012).
Griffith, K. D. et al. In vivo distribution of adoptively transferred indium-111-labeled tumor infiltrating lymphocytes and peripheral blood lymphocytes in patients with metastatic melanoma. J. Natl Cancer Inst. 81, 1709–1717 (1989).
Moon, E. K. et al. Multifactorial T-cell hypofunction that is reversible can limit the efficacy of chimeric antigen receptor-transduced human T cells in solid tumors. Clin. Cancer Res. 20, 4262–4273 (2014).
Moon, E. K. et al. Expression of a functional CCR2 receptor enhances tumor localization and tumor eradication by retargeted human T cells expressing a mesothelin-specific chimeric antibody receptor. Clin. Cancer Res. 17, 4719–4730 (2011).
Caruana, I. et al. Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T lymphocytes. Nat. Med. 21, 524–529 (2015).
Swanton, C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 72, 4875–4882 (2012).
Sotillo, E. et al. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 5, 1282–1295 (2015).
O’Rourke, D. M. et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci. Transl Med. 9, eaaa0984 (2017).
Rapoport, A. P. et al. NY-ESO-1-specific TCR-engineered T cells mediate sustained antigen-specific antitumor effects in myeloma. Nat. Med. 21, 914–921 (2015).
Feng, K. C. et al. Cocktail treatment with EGFR-specific and CD133-specific chimeric antigen receptor-modified T cells in a patient with advanced cholangiocarcinoma. J. Hematol. Oncol. 10, 4 (2017).
Ruella, M. et al. Dual CD19 and CD123 targeting prevents antigen-loss relapses after CD19-directed immunotherapies. J. Clin. Invest. 126, 3814–3826 (2016).
Simon, B., Harrer, D. C., Schuler-Thurner, B., Schuler, G. & Uslu, U. Arming T cells with a gp100-specific TCR and a CSPG4-specific CAR using combined DNA- and RNA-based receptor transfer. Cancers 11, 696 (2019).
Hegde, M. et al. Tandem CAR T cells targeting HER2 and IL13Ralpha2 mitigate tumor antigen escape. J. Clin. Invest. 126, 3036–3052 (2016).
Zah, E. et al. Systematically optimized BCMA/CS1 bispecific CAR-T cells robustly control heterogeneous multiple myeloma. Nat. Commun. 11, 2283 (2020).
de Larrea, C. F. et al. Defining an optimal dual-targeted CAR T-cell therapy approach simultaneously targeting BCMA and GPRC5D to prevent BCMA escape-driven relapse in multiple myeloma. Blood Cancer Discov. 1, 146–154 (2020).
Han, X., Wang, Y., Wei, J. & Han, W. Multi-antigen-targeted chimeric antigen receptor T cells for cancer therapy. J. Hematol. Oncol. 12, 128 (2019).
Bielamowicz, K. et al. Trivalent CAR T cells overcome interpatient antigenic variability in glioblastoma. Neuro Oncol. 20, 506–518 (2018).
Kudo, K. et al. T lymphocytes expressing a CD16 signaling receptor exert antibody-dependent cancer cell killing. Cancer Res. 74, 93–103 (2014).
Landgraf, K. E. et al. ConvertibleCARs: a chimeric antigen receptor system for flexible control of activity and antigen targeting. Commun. Biol. 3, 296 (2020).
Minutolo, N. G., Hollander, E. E. & Powell, D. J., Jr. The emergence of universal immune receptor T cell therapy for cancer. Front. Oncol. 9, 176 (2019).
Minutolo, N. G. et al. Quantitative control of gene-engineered T-cell activity through the covalent attachment of targeting ligands to a universal immune receptor. J. Am. Chem. Soc. 142, 6554–6568 (2020).
Adachi, K. et al. IL-7 and CCL19 expression in CAR-T cells improves immune cell infiltration and CAR-T cell survival in the tumor. Nat. Biotechnol. 36, 346–351 (2018).
Luo, H. et al. Coexpression of IL7 and CCL21 increases efficacy of CAR-T cells in solid tumors without requiring preconditioned lymphodepletion. Clin. Cancer Res. 26, 5494–5505 (2020).
Wang, N. et al. Efficacy and safety of CAR19/22 T-cell cocktail therapy in patients with refractory/relapsed B-cell malignancies. Blood 135, 17–27 (2020).
Yan, Z. et al. A combination of humanised anti-CD19 and anti-BCMA CAR T cells in patients with relapsed or refractory multiple myeloma: a single-arm, phase 2 trial. Lancet Haematol. 6, e521–e529 (2019).
Zhao, W. H. et al. A phase 1, open-label study of LCAR-B38M, a chimeric antigen receptor T cell therapy directed against B cell maturation antigen, in patients with relapsed or refractory multiple myeloma. J. Hematol. Oncol. 11, 141 (2018).
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013).
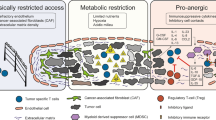
Ecker, C. & Riley, J. L. Translating in vitro T cell metabolic findings to in vivo tumor models of nutrient competition. Cell Metab. 28, 190–195 (2018).
Munn, D. H. & Bronte, V. Immune suppressive mechanisms in the tumor microenvironment. Curr. Opin. Immunol. 39, 1–6 (2016).
Lu, Y. et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 26, 732–740 (2020).
Neuzillet, C. et al. Targeting the TGFbeta pathway for cancer therapy. Pharmacol. Ther. 147, 22–31 (2015).
Kloss, C. C. et al. Dominant-negative TGF-beta receptor enhances PSMA-targeted human CAR T cell proliferation and augments prostate cancer eradication. Mol. Ther. 26, 1855–1866 (2018).
Powell, A. B. et al. Medulloblastoma rendered susceptible to NK-cell attack by TGFbeta neutralization. J. Transl. Med. 17, 321 (2019).
Bollard, C. M. et al. Tumor-specific T-cells engineered to overcome tumor immune evasion induce clinical responses in patients with relapsed hodgkin lymphoma. J. Clin. Oncol. 36, 1128–1139 (2018).
Beavis, P. A. et al. Targeting the adenosine 2A receptor enhances chimeric antigen receptor T cell efficacy. J. Clin. Invest. 127, 929–941 (2017).
Sim, G. C. et al. IL-2 therapy promotes suppressive ICOS+ Treg expansion in melanoma patients. J. Clin. Invest. 124, 99–110 (2014).
Suryadevara, C. M. et al. Preventing Lck activation in CAR T cells confers Treg resistance but requires 4-1BB signaling for them to persist and treat solid tumors in nonlymphodepleted hosts. Clin. Cancer Res. 25, 358–368 (2019).
Franchina, D. G., He, F. & Brenner, D. Survival of the fittest: cancer challenges T cell metabolism. Cancer Lett. 412, 216–223 (2018).
Kawalekar, O. U. et al. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T Cells. Immunity 44, 712 (2016).
Fultang, L. et al. Metabolic engineering against the arginine microenvironment enhances CAR-T cell proliferation and therapeutic activity. Blood 136, 1155–1160 (2020).
Leonard, J. P. et al. Effects of single-dose interleukin-12 exposure on interleukin-12-associated toxicity and interferon-gamma production. Blood 90, 2541–2548 (1997).
Zhang, L. et al. Improving adoptive T cell therapy by targeting and controlling IL-12 expression to the tumor environment. Mol. Ther. 19, 751–759 (2011).
Joung, J. et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
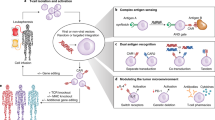
Shifrut, E. et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell 175, 1958–1971.e15 (2018). This study uses a genome-wide CRISPR screen to unearth proteins that limit T cell proliferation and killing in the tumour microenvironment.
Cortez, J. T. et al. CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature 582, 416–420 (2020).
Dong, M. B. et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell 178, 1189–1204.e23 (2019).
Roth, T. L. et al. Pooled knockin targeting for genome engineering of cellular immunotherapies. Cell 181, 728–744.e21 (2020).
Wei, J. et al. PD-1 silencing impairs the anti-tumor function of chimeric antigen receptor modified T cells by inhibiting proliferation activity. J. Immunother. Cancer 7, 209 (2019).
Cherkassky, L. et al. Human CAR T cells with cell-intrinsic PD-1 checkpoint blockade resist tumor-mediated inhibition. J. Clin. Invest. 126, 3130–3144 (2016).
Ankri, C., Shamalov, K., Horovitz-Fried, M., Mauer, S. & Cohen, C. J. Human T cells engineered to express a programmed death 1/28 costimulatory retargeting molecule display enhanced antitumor activity. J. Immunol. 191, 4121–4129 (2013).
Parry, R. V. et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol. Cell Biol. 25, 9543–9553 (2005).
Rafiq, S. et al. Targeted delivery of a PD-1-blocking scFv by CAR-T cells enhances anti-tumor efficacy in vivo. Nat. Biotechnol. 36, 847–856 (2018).
Zhang, A. et al. Secretion of human soluble programmed cell death protein 1 by chimeric antigen receptor-modified T cells enhances anti-tumor efficacy. Cytotherapy 22, 734–743 (2020).
Chang, Z. L., Silver, P. A. & Chen, Y. Y. Identification and selective expansion of functionally superior T cells expressing chimeric antigen receptors. J. Transl. Med. 13, 161 (2015).
Khan, O. et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218 (2019).
Chen, Z. et al. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity 51, 840–855.e5 (2019).
Chen, J. et al. NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534 (2019).
Odorizzi, P. M., Pauken, K. E., Paley, M. A., Sharpe, A. & Wherry, E. J. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J. Exp. Med. 212, 1125–1137 (2015).
Greenbaum, U. et al. Chimeric antigen receptor T-cell therapy toxicities. Br. J. Clin. Pharmacol. https://doi.org/10.1111/bcp.14403 (2020).
Curran, K. J. et al. Toxicity and response after CD19-specific CAR T-cell therapy in pediatric/young adult relapsed/refractory B-ALL. Blood 134, 2361–2368 (2019).
Neelapu, S. S. et al. Chimeric antigen receptor T-cell therapy - assessment and management of toxicities. Nat. Rev. Clin. Oncol. 15, 47–62 (2018).
Howard, S. C., Trifilio, S., Gregory, T. K., Baxter, N. & McBride, A. Tumor lysis syndrome in the era of novel and targeted agents in patients with hematologic malignancies: a systematic review. Ann. Hematol. 95, 563–573 (2016).
Fried, S. et al. Early and late hematologic toxicity following CD19 CAR-T cells. Bone Marrow Transpl. 54, 1643–1650 (2019).
Teachey, D. T. et al. Identification of predictive biomarkers for cytokine release syndrome after chimeric antigen receptor T-cell therapy for acute lymphoblastic leukemia. Cancer Discov. 6, 664–679 (2016).
Ruella, M. et al. Induction of resistance to chimeric antigen receptor T cell therapy by transduction of a single leukemic B cell. Nat. Med. 24, 1499–1503 (2018).
Hacein-Bey-Abina, S. et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 302, 415–419 (2003).
Marcucci, K. T. et al. Retroviral and lentiviral safety analysis of gene-modified T cell products and infused HIV and oncology patients. Mol. Ther. 26, 269–279 (2018).
Morgan, R. A. et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J. Immunother. 36, 133–151 (2013).
Beatty, G. L. et al. Activity of mesothelin-specific chimeric antigen receptor T cells against pancreatic carcinoma metastases in a phase 1 trial. Gastroenterology 155, 29–32 (2018).
Maus, M. V. et al. T cells expressing chimeric antigen receptors can cause anaphylaxis in humans. Cancer Immunol. Res. 1, 26–31 (2013).
Tasian, S. K. et al. Optimized depletion of chimeric antigen receptor T cells in murine xenograft models of human acute myeloid leukemia. Blood 129, 2395–2407 (2017).
Jonnalagadda, M. et al. Efficient selection of genetically modified human T cells using methotrexate-resistant human dihydrofolate reductase. Gene Ther. 20, 853–860 (2013).
Cavaco, M., Gaspar, D., Arb Castanho, M. & Neves, V. Antibodies for the treatment of brain metastases, a dream or a reality? Pharmaceutics 12, 62 (2020).
Gargett, T. & Brown, M. P. The inducible caspase-9 suicide gene system as a “safety switch” to limit on-target, off-tumor toxicities of chimeric antigen receptor T cells. Front. Pharmacol. 5, 235 (2014).
Gu, X., He, D., Li, C., Wang, H. & Yang, G. Development of inducible CD19-CAR T cells with a Tet-on system for controlled activity and enhanced clinical safety. Int. J. Mol. Sci. 19, 3455 (2018).
Drent, E. et al. Feasibility of controlling CD38-CAR T cell activity with a Tet-on inducible CAR design. PLoS ONE 13, e0197349 (2018).
Sakemura, R. et al. A Tet-on inducible system for controlling CD19-chimeric antigen receptor expression upon drug administration. Cancer Immunol. Res. 4, 658–668 (2016).
Zhang, R. Y. et al. Doxycycline inducible chimeric antigen receptor T cells targeting CD147 for hepatocellular carcinoma therapy. Front. Cell Dev. Biol. 7, 233 (2019).
Foster, A. E. et al. Regulated expansion and survival of chimeric antigen receptor-modified T cells using small molecule-dependent inducible MyD88/CD40. Mol. Ther. 25, 2176–2188 (2017).
Duong, M. T. et al. Two-dimensional regulation of CAR-T cell therapy with orthogonal switches. Mol. Ther. Oncolytics 12, 124–137 (2019).
Wu, C. Y., Roybal, K. T., Puchner, E. M., Onuffer, J. & Lim, W. A. Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science 350, aab4077 (2015).
Huang, Z. et al. Engineering light-controllable CAR T cells for cancer immunotherapy. Sci. Adv. 6, eaay9209 (2020).
Lee, S. M. et al. A Chemical switch system to modulate chimeric antigen receptor T cell activity through proteolysis-targeting chimaera technology. ACS Synth. Biol. 9, 987–992 (2020).
Juillerat, A. et al. Modulation of chimeric antigen receptor surface expression by a small molecule switch. BMC Biotechnol. 19, 44 (2019).
Richman, S. A. et al. Ligand-induced degradation of a CAR permits reversible remote control of CAR T cell activity in vitro and in vivo. Mol. Ther. 28, 1600–1613 (2020).
Mestermann, K. et al. The tyrosine kinase inhibitor dasatinib acts as a pharmacologic on/off switch for CAR T cells. Sci. Transl Med. 11, eaau5907 (2019). This article reports a pharmacological approach to modulating T cell function in vivo.
Ebert, L. M., Yu, W., Gargett, T. & Brown, M. P. Logic-gated approaches to extend the utility of chimeric antigen receptor T-cell technology. Biochem. Soc. Trans. 46, 391–401 (2018).
Zah, E., Lin, M. Y., Silva-Benedict, A., Jensen, M. C. & Chen, Y. Y. T cells expressing CD19/CD20 bispecific chimeric antigen receptors prevent antigen escape by malignant B Cells. Cancer Immunol. Res. 4, 498–508 (2016).
Fedorov, V. D., Themeli, M. & Sadelain, M. PD-1- and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci. Transl Med. 5, 215ra172 (2013).
Lajoie, M. J. et al. Designed protein logic to target cells with precise combinations of surface antigens. Science 369, 1637–1643 (2020).
Srivastava, S. et al. Logic-gated ROR1 chimeric antigen receptor expression rescues T cell-mediated toxicity to normal tissues and enables selective tumor targeting. Cancer Cell 35, 489–503 e488 (2019).
Roybal, K. T. et al. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell 164, 770–779 (2016). This study describes the development of synNotch CAR T cells operating on IF/THEN logic.
Juillerat, A. et al. An oxygen sensitive self-decision making engineered CAR T-cell. Sci. Rep. 7, 39833 (2017).
Michaels, Y. S. et al. Precise tuning of gene expression levels in mammalian cells. Nat. Commun. 10, 818 (2019).
Chassin, H. et al. A modular degron library for synthetic circuits in mammalian cells. Nat. Commun. 10, 2013 (2019).
Ying, Z. et al. A safe and potent anti-CD19 CAR T cell therapy. Nat. Med. 25, 947–953 (2019).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). This study describes the development of prime editing for genome editing in the absence of DNA double-strand breaks.
Tebas, P. et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 370, 901–910 (2014).
Paschon, D. E. et al. Diversifying the structure of zinc finger nucleases for high-precision genome editing. Nat. Commun. 10, 1133 (2019).
Perez, E. E. et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 26, 808–816 (2008).
Wilen, C. B. et al. Engineering HIV-resistant human CD4+ T cells with CXCR4-specific zinc-finger nucleases. PLoS Pathog. 7, e1002020 (2011).
Yuan, J. et al. Zinc-finger nuclease editing of human cxcr4 promotes HIV-1 CD4+ T cell resistance and enrichment. Mol. Ther. 20, 849–859 (2012).
Provasi, E. et al. Editing T cell specificity towards leukemia by zinc finger nucleases and lentiviral gene transfer. Nat. Med. 18, 807–815 (2012).
Qasim, W. et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci. Transl Med. 9, eaaj2013 (2017).
Valton, J. et al. A multidrug-resistant engineered CAR T cell for allogeneic combination immunotherapy. Mol. Ther. 23, 1507–1518 (2015).
Mussolino, C. et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 39, 9283–9293 (2011).
Sachdeva, M. et al. Repurposing endogenous immune pathways to tailor and control chimeric antigen receptor T cell functionality. Nat. Commun. 10, 5100 (2019).
Ren, J. et al. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin. Cancer Res. 23, 2255–2266 (2017). The authors describe a CRISPR–Cas9-based triple knockout of genes encoding TCR, β2M and PD1 in primary human T cells.
Liu, Z. et al. Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4+ T cells from HIV-1 infection. Cell Biosci. 7, 47 (2017).
Anderson, W., Thorpe, J., Long, S. A. & Rawlings, D. J. Efficient CRISPR/Cas9 disruption of autoimmune-associated genes reveals key signaling programs in primary human T cells. J. Immunol. 203, 3166–3178 (2019).
Gaudelli, N. M. et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 38, 892–900 (2020).
Gornalusse, G. G. et al. HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat. Biotechnol. 35, 765–772 (2017).
Mo, F. et al. Engineered off-the-shelf therapeutic T cells resist host immune rejection. Nat. Biotechnol. (2020).
Wu, W. et al. Multiple signaling roles of CD3epsilon and its application in CAR-T cell therapy. Cell 182, 855–871.e23 (2020).
Roberts, M. R. et al. Targeting of human immunodeficiency virus-infected cells by CD8+ T lymphocytes armed with universal T-cell receptors. Blood 84, 2878–2889 (1994).
Maher, J., Brentjens, R. J., Gunset, G., Riviere, I. & Sadelain, M. Human T-lymphocyte cytotoxicity and proliferation directed by a single chimeric TCRzeta /CD28 receptor. Nat. Biotechnol. 20, 70–75 (2002).
Porter, D. L., Levine, B. L., Kalos, M., Bagg, A. & June, C. H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 365, 725–733 (2011).
Kochenderfer, J. N. et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood 119, 2709–2720 (2012).
Brentjens, R. J. et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci. Transl Med. 5, 177ra138 (2013).
Porter, D. L. et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci. Transl Med. 7, 303ra139 (2015).
Kim, G. B., Hege, K. & Riley, J. L. CAR talk: how cancer-specific CAR T cells can instruct how to build CAR T cells to cure HIV. Front. Immunol. 10, 2310 (2019).
Maldini, C. R. et al. HIV-resistant and HIV-specific CAR-modified CD4+ T cells mitigate HIV disease progression and confer CD4+ T cell help in vivo. Mol. Ther. 28, 1585–1599 (2020).
Herzig, E. et al. Attacking latent HIV with convertible CAR-T cells, a highly adaptable killing platform. Cell 179, 880–894.e10 (2019).
Anthony-Gonda, K. et al. Multispecific anti-HIV duoCAR-T cells display broad in vitro antiviral activity and potent in vivo elimination of HIV-infected cells in a humanized mouse model. Sci. Transl Med. 11, eaav5685 (2019).
Ali, A. et al. HIV-1-specific chimeric antigen receptors based on broadly neutralizing antibodies. J. Virol. 90, 6999–7006 (2016).
Festag, M. M. et al. Evaluation of a fully human, hepatitis b virus-specific chimeric antigen receptor in an immunocompetent mouse model. Mol. Ther. 27, 947–959 (2019).
Sautto, G. A. et al. Chimeric antigen receptor (CAR)-engineered T cells redirected against hepatitis C virus (HCV) E2 glycoprotein. Gut 65, 512–523 (2016).
Kumaresan, P. R. et al. Bioengineering T cells to target carbohydrate to treat opportunistic fungal infection. Proc. Natl Acad. Sci. USA 111, 10660–10665 (2014).
Bitinaite, J., Wah, D. A., Aggarwal, A. K. & Schildkraut, I. FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA 95, 10570–10575 (1998).
Kim, Y. et al. A library of TAL effector nucleases spanning the human genome. Nat. Biotechnol. 31, 251–258 (2013).
Boissel, S. et al. megaTALs: a rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 42, 2591–2601 (2014).
Osborn, M. J. et al. Evaluation of TCR gene editing achieved by TALENs, CRISPR/Cas9, and megaTAL nucleases. Mol. Ther. 24, 570–581 (2016).
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Konermann, S. et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 (2013).
Yeo, N. C. et al. An enhanced CRISPR repressor for targeted mammalian gene regulation. Nat. Methods 15, 611–616 (2018).
Huang, Y. H. et al. DNA epigenome editing using CRISPR-Cas SunTag-directed DNMT3A. Genome Biol. 18, 176 (2017).
Koonin, E. V., Makarova, K. S. & Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 37, 67–78 (2017).
Komor, A. C., Badran, A. H. & Liu, D. R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 (2017).
Acknowledgements
The authors are grateful to the members of the Center for Cellular Immunotherapies for in-depth discussions regarding the topics discussed in this Review and apologize to the many investigators whose work they were unable to cite. The authors gratefully acknowledge funding from the NIH (U19AI117950, U19AI149680 UM1AI126620 and UG3DK122644), Helmsley Charitable Trust and Tmunity Therapeutics.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
J.L.R. is a co-founder and shareholder of Tmunity Therapeutics. He has been in receipt of research funding from Tmunity Therapeutics. N.C.S. is a shareholder of Tmunity Therapeutics and Fate Therapeutics. G.I.E. declares no competing interests.
Additional information
Peer review information
Nature Reviews Genetics thanks M. K. Levings, Z. Li, E. L. Smith, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
NCT01236573: https://clinicaltrials.gov/ct2/show/NCT01236573
NCT03240328: https://clinicaltrials.gov/ct2/show/NCT03240328
NCT03617198: https://clinicaltrials.gov/ct2/show/NCT03617198
NCT04422912: https://clinicaltrials.gov/ct2/show/NCT04422912
Glossary
- Allogeneic
-
A term to denote a genetically different individual of the same species.
- Antigen loss
-
A mechanism of cancer or pathogen recurrence in which the target (tumour or pathogen) escapes engineered T cell recognition, typically via exon skipping, downregulation or alteration of the target antigen.
- Epitope spreading
-
Diversification of the immune response by endogenous immune cells against new targets following engineered T cell therapy.
- Boolean logic gate
-
AND, OR and NOT functions, which can describe the response of combinations of immune receptors to multiple antigens.
- Immune synapse
-
The location of physical interaction between an immune cell and its activator, either another immune cell or a non-haematopoietic target cell. The strength and the type of signals exchanged at the immune synapse can modulate an immune response.
- Single-chain variable fragment
-
(scFv). A linear transposition of an antibody’s heavy and light chains separated by a flexible linker, which retains the antigen binding capacity of the original antibody and can be used as the binder domain of a chimeric antigen receptor molecule.
- Monoclonal antibodies
-
(mAbs). Antibodies purified from a single B cell hybridoma that can be used as a diagnostic or therapeutic product.
- Fc-dependent mechanisms
-
A series of effector modalities by which targets bound by antibodies are depleted via recognition of the constant region of the antibody molecule.
Rights and permissions
About this article
Cite this article
Ellis, G.I., Sheppard, N.C. & Riley, J.L. Genetic engineering of T cells for immunotherapy. Nat Rev Genet 22, 427–447 (2021). https://doi.org/10.1038/s41576-021-00329-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-021-00329-9
This article is cited by
-
Hydrophobic mismatch drives self-organization of designer proteins into synthetic membranes
Nature Communications (2024)
-
Programmable synthetic receptors: the next-generation of cell and gene therapies
Signal Transduction and Targeted Therapy (2024)
-
The screening, identification, design and clinical application of tumor-specific neoantigens for TCR-T cells
Molecular Cancer (2023)
-
CAR T therapy beyond cancer: the evolution of a living drug
Nature (2023)
-
Host-directed immunotherapy of viral and bacterial infections: past, present and future
Nature Reviews Immunology (2023)