Future Perspective
The application of pharmacogenomics to complex diseases characterized by a great phenotypic and genetic variability might constitute an almost unconquerable challenge. The phenotype of response is a complex and heterogeneous trait by itself, and it is definable as a continuous more than a dichotomous (response/nonresponse) trait. It has been stated that, given the weak effects of susceptibility genotypes, it is theoretically improbable that genetic screening will be used for the assessment of risk or prognosis in complex diseases.[158]
Although this evidence may lead us to conclude that translating pharmacogenomic findings to personalized medicine in complex diseases will be difficult, some strategies can be employed to overcome the challenges and lay the foundation for pharmacogenomic implementation in clinics. For instance, in complex diseases, missing heritability might be ascribed to the presence of rare variants and structural variations.[159] It has been stated that DNA sequencing might allow the identification of rare SNPs either in target regions or in the whole genome. The use of subjects at the extreme of a quantitative trait, such as complete nonresponse versus response to specific treatment, has been suggested as a useful mechanism for the identification of associated variants, both rare and common, by sequencing.[159] Moreover, some analytical approaches, such as case-only genome-wide interaction, can provide a straightforward method for detecting genetic interactions related to treatment response in complex diseases.[160] On the whole, this evidence might provide a framework of operational and decisional criteria for setting the course leading to widespread use of pharmacogenomics in public health.
In addition, novel concepts emerge in the burgeoning field of pharmacogenomics, for example, in the area of antibiotic therapy, where sequence variations in the mitochondrial DNA (mtDNA) can constitute putatively useful pharmacogenetic markers.[161] Cell systems, known as transmitochondrial cell lines or cybrids, where the mtDNA of a parental cell line is depleted and the resulting cell is fused to enucleated cells, have been employed to analyze the interaction between antibiotics and mtDNA genetic variants. These cell lines have the same nuclear genetic composition and grow in the same environment, while only differing in their mtDNA, which would yield distinct phenotypic differences as a result of drug treatment.[162,163] Such cell lines have been used to correlate human chloramphenicol resistance with two mtDNA variations, namely m.2939C>A and m.2991T>C in the MT-RNR2 gene.[162,163] This experimental system has been also used to explore susceptibility to erythromycin.[164] Such approaches would give us a new perspective of the pharmacogenomics of antibiotic therapy, and could possibly assist towards optimizing or increasing the number of available antibiotics on the basis of the patient's genetic background.
Furthermore, pharmacogenomics can be associated with variable response to drug therapy in organ transplantation. For example, tacrolimus is perhaps the best established example of the CYP3A5 gene effect on drug disposition and dosage, with the CYP3A5*3 allele correlating with lower dose of tacrolimus to achieve therapeutic blood concentrations. This same observation has been made for every type of solid organ transplant, but unfortunately this effect is not uniform among patients, especially in those bearing the CYP3A5*3/*3 genotype. The effectiveness of other drugs used in organ transplantation have been correlated vis à vis to a number of gene alleles, such as azathioprine with TPMT alleles, ciclosporin with ABCB1 alleles, sirolimus, like tacrolimus, with CYP3A5 alleles, corticosteroids with ABCB1 and IL-10 polymorphisms (reviewed in[165]).
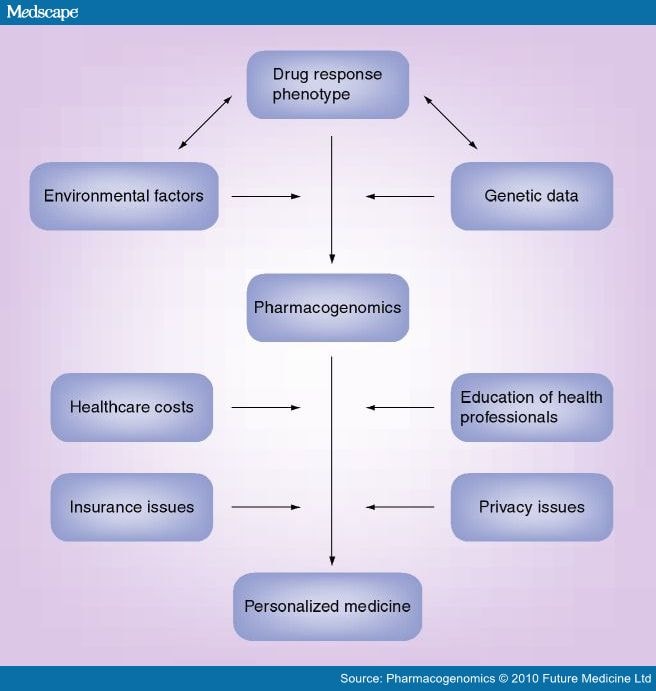
Besides the promising evidence towards a wider adoption of pharmacogenomics in the clinical settings, a number of obstacles on its full implementation for the achievement of personalized therapy must be taken into account (Figure 1). In a broad perspective, personalized medicine will require changes in healthcare infrastructure, diagnostic business models and a reimbursement policy on the part of government and private payers. To address this need, the Personalized Medicine Coalition (PMC; Washington, DC, USA) was founded as a nonprofit organization of pharmaceutical biotechnology, diagnostic and information technology companies, healthcare providers and payers, patient advocacy groups, industry policy organizations, academic institutions and government agencies.[166] PMC attempts to facilitate the use of molecular diagnostics and personalized medicine approaches, providing opinion leadership, help in training the public, and conveying information to the media, government officials and healthcare leaders. Other initiatives by nonprofit foundations were undertaken in the last few years to facilitate the implementation of personalized medicine in everyday healthcare. In October 2009, leaders from the field of research, medicine, industry, government and philanthropy founded the Mayflower Action Group Initiative, an idea instigated by Brain Research And Integrative Neuroscience Network (BRAINNet), a new nonprofit foundation that provides a database on the human brain using standardized methods.[167] The foundation aims to address the need for combining genetic information, electrical measurements of brain and body function, structural and functional MRI, and cognitive and medical history data within a single framework. These data are from both healthy people and those experiencing a range of brain-related illnesses, and are freely provided for research and scientific publication, thereby maximizing and sharing benefits. Furthermore, the Pharmacogenetics for Every Nation Initiative (PGENI[210]) is a worldwide initiative with the stated goal of developing "…innovative strategies for Health Bodies to integrate pharmacogenetics into public health decision-making without placing an extra burden on sparse healthcare funds". Such efforts would be particularly useful for developing nations to defray healthcare costs and improve quality of life by minimizing ADRs.
Figure 1.
The discovery of pharmacogenomic determinants able to predict treatment outcome in specific populations of patients can be represented as a heuristic process that takes into account different empirical factors acting at a phenotypic, genetic and environmental level.
Specifically, a refined phenotype of treatment response is needed in order to empower the pharmacogenomic approach. The drug-response phenotype can result from the complex interplay with environmental and genomic factors. Once identified, pharmacogenomic predictors can enter a clinical implementation pipeline leading to their practical application into personalized therapy. This path might present different degrees of efficacy, depending on the influence of factors such as privacy and insurance issues, healthcare costs and the educational level of healthcare providers. Actions undertaken at these levels can facilitate the process of clinical implementation, consequently leading to empowered personalized medicine tools.
The era of personalized medicine has already begun, and even though it is not yet a widespread practice, ongoing international efforts confirm that the concept of personalized prescription is close to becoming a reality.
While pharmacogenomics in psychiatry is a rapidly emerging field, it has to face difficulties related to the complexity of the phenotype and the polygenic features of response to psychotropic medication. Nevertheless, promising results from genome-wide studies on response to lithium and antidepressants currently underway might provide validation of the candidate genes mentioned earlier, as well as identification of novel genes. Application of sophisticated 'machine-learning' algorithms to data from genome-wide studies of treatment response will allow researchers to develop personalized treatments based on genetic data.
In light of these assumptions, it is reasonable to presume that the implementation of genetic data for the design of a personalized prescription will be achieved more quickly in other fields such as oncology, where healthcare professionals have access to tissues permitting various types of testing. However, increasing pharmacological information on the genetic bases of response to medications, drug–drug interactions and variability due to clinical and environmental factors will lead to more widespread use of pharmacogenomics in psychiatry in the near future.
Personalized medicine has always been a component of good medical practice. Genetic tests may provide new tools, but do not change the fundamental goal of clinicians: to adapt available medical tests and technologies to the individual circumstances of their patients. As genetic tests become widely available, personalized medicine will make wise use of genetic information in analyzing the complex picture regarding variability in response to medication.
Acknowledgements
We are grateful to Tarryn Greenberg, Managing Commissioning Editor (Pharmacogenomics), for soliciting this article, parts of which have been presented at the 2009 Golden Helix Symposium® 'Pharmacogenomics: Paving the path to personalized medicine', held in Athens, Greece, 15–17 October 2009. The authors gratefully acknowledge Ms Mary Groeneweg for commenting on the manuscript and Mr Sean Scaccia for his assistance with the graphical design.
Financial & competing interests disclosure
This work was partly supported by the Regional Councillorship of Health, 'Regione Autonoma della Sardegna' with a grant dedicated to Drug Information and surveillance Projects to Alessio Squassina, Mirko Manchia, Maria Del Zompo and by grants from the European Commission (GEN2PHEN; FP7200754 and the Research Promotion Foundation of Cyprus (ΠΔE046_02) and the University of Patras Research budget to George P Patrinos. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Pharmacogenomics. 2010;11(8):1149-1167. © 2010 Future Medicine Ltd.
Cite this: Realities and Expectations of Pharmacogenomics and Personalized Medicine: Impact of Translating Genetic Knowledge into Clinical Practice - Medscape - Aug 01, 2010.