DNA SOS: Understanding DNA signaling pathways
When cells suffer DNA damage, they send out an SOS signal. When the repair crew arrives, the emergency signal is cancelled as it is no longer needed. This two-stage process is an important one in many different diseases, but the underlying mechanisms are not well understood. ADP-ribosylation is one of the DNA damage response mechanisms used by cells to send out an SOS signal; and the ARH family of enzymes is involved in canceling the signal. In work recently published in Cell Chemical Biology, researchers from the University of Oxford's DNA Damage Response Lab investigated the structure of two ADP-(ribosyl)hydrolases (ARHs). Their results provide important insights into the differences between these two proteins and offer useful tools for targeted drug design.
DNA repair
The DNA in our cells is under constant attack, and estimates suggest it is damaged thousands of times every day. Left untreated, these lesions can lead to mutations, cell death, or—in the worst case—cancer. However, cells have a number of DNA damage response pathways by which they can counteract DNA damage, and these are generally sufficient to maintain a healthy genome.
Previous research has shown that cancer, neurodegenerative diseases, immunodeficiency or developmental abnormalities are linked to defects in DNA damage response pathways. The University of Oxford's DNA Damage Response Lab studies the protein functions and pathways underpinning genome stability, to increase our understanding and help to develop new ways to prevent and treat disease.
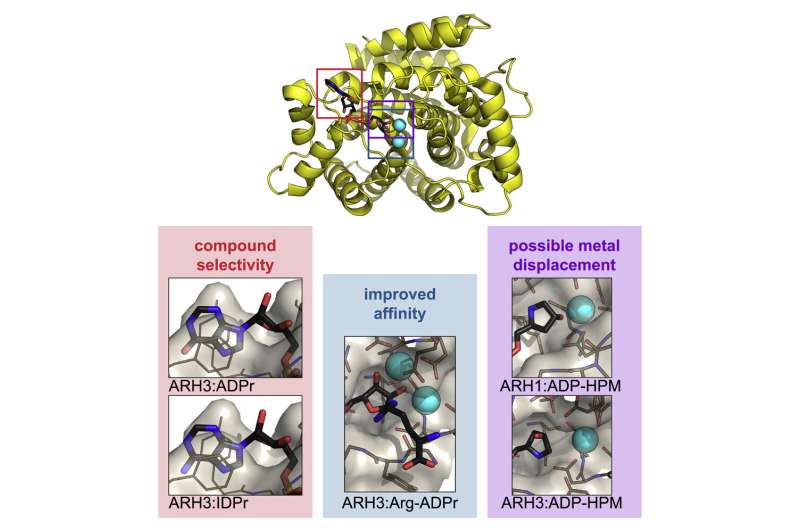
In this new work, researchers used Diamond's macromolecular crystallography (MX) beamlines to investigate the structure of ARH1 and ARH3. ARH1 is essential in tumor suppression and plays a role in defending against bacterial toxins. ARH3 is known to be involved in the DNA damage response.
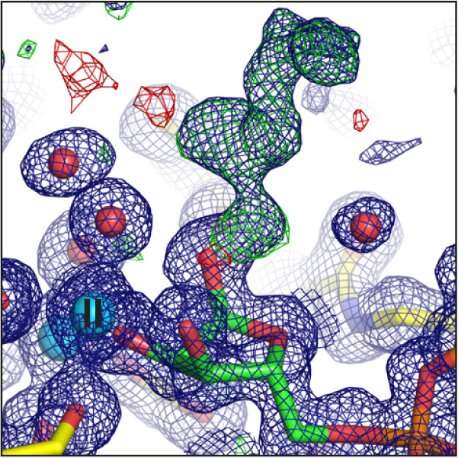
Crystallising human ARH3 is challenging, as the protein will only crystallize in the presence of a ligand, which cannot always be detected in the corresponding structure. In order to circumvent this problem, Dr. Johannes Rack used a homologue of ARH3 from the West Indian Ocean coelacanth (Latimeria chalumnae), which crystallized more readily. This allowed the researchers to solve crystal structures with and without a number of ligands that they could not obtain with the human protein. These also turned out to be the first ever coelacanth protein structures published.
MX at Diamond
Experiments for this research were carried out on beamlines I03, I04, I04-1, and I24. The ARH proteins are magnesium-dependent, and their structure had to be determined both with and without magnesium present. The paper, therefore, contains ten protein structures, collected at atomic resolution. This kind of intensive research was possible because the University of Oxford has BAG access to Diamond, which allows groups of users to fill a beamtime shift with a series of short experiments.
Dr. Antonio Ariza explains: "Before Diamond opened, I had to fly to Grenoble to conduct MX experiments, which was a considerable time investment. Experiments are so quick now, and Diamond makes continuous improvements. I visit Diamond several times every month now. Many of our experiments could be automated, but I like coming in person to get my hands dirty, and to keep up with the changes."
ARH1 and ARH3 are highly specialized enzymes, catalyzing different reactions. However, the results of these experiments show that they are structurally very similar, particularly in terms of their active sites. The new data on these structures will provide insights into their operation, and help us understand their differences. With no known inhibitors for this family of enzymes, the researchers hope that their work will inform future drug discovery. Their findings provide opportunities to design compounds that modulate the activities of ARH enzymes, with great promise for therapeutic potential.
Experiments for this research were carried out on beamlines I03, I04, I04-1, and I24.
More information: Johannes Gregor Matthias Rack et al. (ADP-ribosyl)hydrolases: Structural Basis for Differential Substrate Recognition and Inhibition, Cell Chemical Biology (2018). DOI: 10.1016/j.chembiol.2018.11.001
Journal information: Cell Chemical Biology
Provided by Diamond Light Source