Two-step process identifies bacterial infection quickly to help combat antibiotic resistance
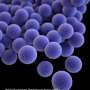
Bacterial infections are a major cause of death in children and adults worldwide. Without effective antibiotics, patients deteriorate rapidly and can die within hours. However, the standard process to identify the bacteria causing an infection and match them with the best antibiotic treatment can take days.
A research team from the Harry Perkins Institute of Medical Research, The University of Western Australia and PathWest Laboratory Medicine WA including Perth renal physician, Dr. Aron Chakera and UWA Forrest Prospect Fellow, Dr. Kieran Mulroney, have developed a faster, more effective process to confirm infections and find the most effective treatment for them.
"The established method involves growing bacteria from a patient sample then applying different antibiotics to see which are effective. Patients with serious infections cannot wait the several days it can take to return antibiotic test results. Consequently, the patient's doctor has to rely on a best guess, 'one-size-fits-all,' antibiotic choice to treat patients.
"The biggest problem with prescribing broad-spectrum antibiotics is that it encourages some bacteria to become resistant to the antibiotics. This is a growing and serious problem world-wide, because antibiotic resistant bacteria can spread from person to person and reduce treatment options.
"The overuse of broad-spectrum antibiotics is one of the key drivers in the spread of resistance to antibiotics," Dr. Mulroney said.
Quicker, more accurate, tests mean treatment can be targeted to each infection, so the antibiotics are just right—not too strong, and not ineffective.
"New tests are urgently needed that give doctors evidence they can rely on to select the right antibiotic" he said.
Dr. Mulroney said the team has developed a two-step process:
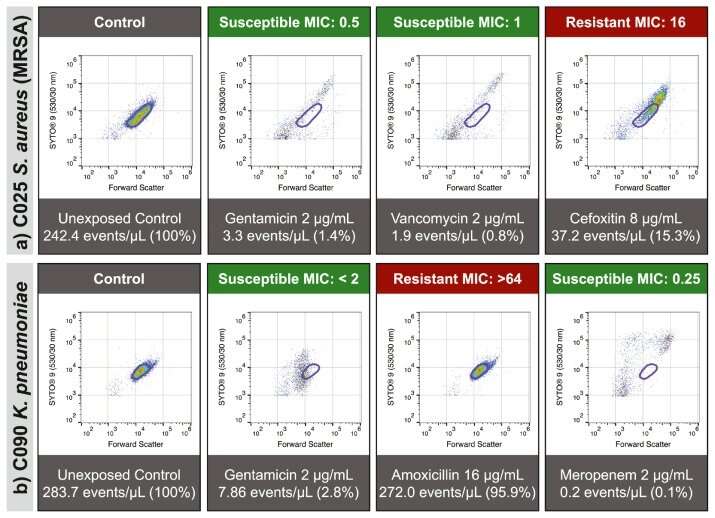
"First, we developed a test to confirm if the cause of the patient's serious illness is a bacterial infection. This test takes 30 minutes, rather than 1–2 days.
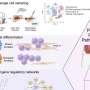
"Once a patient has a confirmed bacterial infection, we then expose the bacteria to different types of antibiotics in the laboratory. Using a device that measures hundreds of thousands of individual bacteria in just a few seconds, the research team can detect the damage antibiotics cause to bacteria, and then use this information to confirm which antibiotic will be an effective treatment. We can predict which antibiotics will be effective to treat that infection with 96.9% accuracy," Dr. Mulroney said.
Sir Charles Gairdner Hospital renal physician Dr. Chakera says for patients with chronic illnesses the discovery will be potentially life-saving.
"As a renal physician I treat patients with end-stage kidney disease who need to be in hospitals or clinics for several hours a week connected to dialysis machines. Many could manage their own dialysis using a surgically implanted catheter, which actually has better outcomes, is far less costly and is more satisfying for patients, but the ever-present fear of infection from the catheter deters many from choosing it.
"This new test would give confidence to patients and their treating doctors," Dr. Chakera said.
WA Country Health Service Translation Fellow, Dr. Tim Inglis has seen the method develop from its earliest test tube observations into a robust clinical laboratory method with a broad range of applications.
"The time and effort it takes to produce accurate antibiotic test results make this technique very attractive to busy clinical laboratories. The last two years has shown us how important test results are in the battle against infection. When the COVID pandemic finally tails off, the challenge of antibiotic resistance will still be there, demanding urgent attention from the clinical lab."
"Even in the most advanced health systems, hospital patients risk bacterial infection through trauma wounds, surgery sites, breathing machines and indwelling catheters. This can lead to pneumonia, urinary tract, abdominal and bloodstream infections. Applying the research team's new technology to these infections is expected to transform how quickly and effectively we treat patients in Western Australia and further afield," Dr. Inglis said.
The study is published in eBioMedicine.
More information: Kieran Mulroney et al, Same-day confirmation of infection and antimicrobial susceptibility profiling using flow cytometry, eBioMedicine (2022). DOI: 10.1016/j.ebiom.2022.104145