Further insights into the SARS-CoV-2 spike protein glycan shield
An unprecedented wealth of information has been gathered on SARS-CoV-2 in a very short period. This information is primarily focused on the cellular entry process and mechanism of antibody recognition where mainly protein-protein interactions occur.
The groups of Dr. Abrescia and Dr. Jiménez-Osés at CIC bioGUNE have combined high-resolution cryo-electron microscopy and computer simulation to understand the correlation between sugar identity and flexibility in the SARS-CoV-2 spike glycoprotein and published the results in Frontiers in Microbiology.
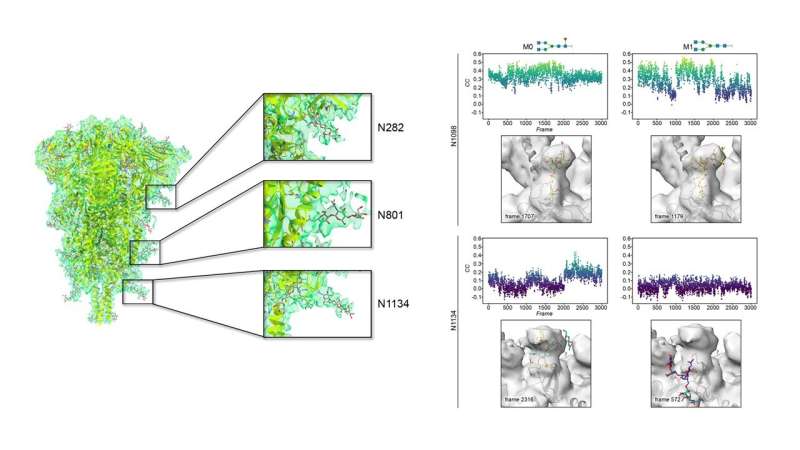
Rapid and free access to high-end Krios microscopes at eBIC-Diamond LS (UK) by the Abrescia Lab has allowed a 3D reconstruction of the spike protein at 4.1 Å resolution with a very limited number of contributing particles (~23,000) in which the density for the decorating glycans is as clear as other maps at higher resolution for which hundreds of thousands of particles were necessary.
The most ordered sugars are located at the so-called S2 domain, which is proximal to the viral membrane. Chemical variations of those glycans discovered by mass spectrometry were modeled on representative glycosylated amino acids by the Jiménez-Osés lab, and showed no significant influence on either protein shielding or glycan flexibility. Mathematical methods were used to compare the cryo-EM density and the time-resolved full-atom computer models. The best fits between the two techniques are characterized by glycans showing very conserved geometries around crucial glycosidic bonds near to the protein.
Being able to predict glycan behavior is relevant because this S2 location on the spike is the ideal target for a pan-ligand capable of neutralizing the virus after cell-entry. This study—also the result of collaborations with Jimenez-Barbero, Millet and Connell labs—shows that experimental and computational tools combined can provide valuable insights on the conformational preferences of inherently flexible and complex glycoconjugates, advancing the discovery of new drugs able to evade the glycan shield of infectious viral proteins.
More information: Soledad Stagnoli et al, Assessing the Mobility of Severe Acute Respiratory Syndrome Coronavirus-2 Spike Protein Glycans by Structural and Computational Methods, Frontiers in Microbiology (2022). DOI: 10.3389/fmicb.2022.870938
Journal information: Frontiers in Microbiology
Provided by CIC bioGUNE





















