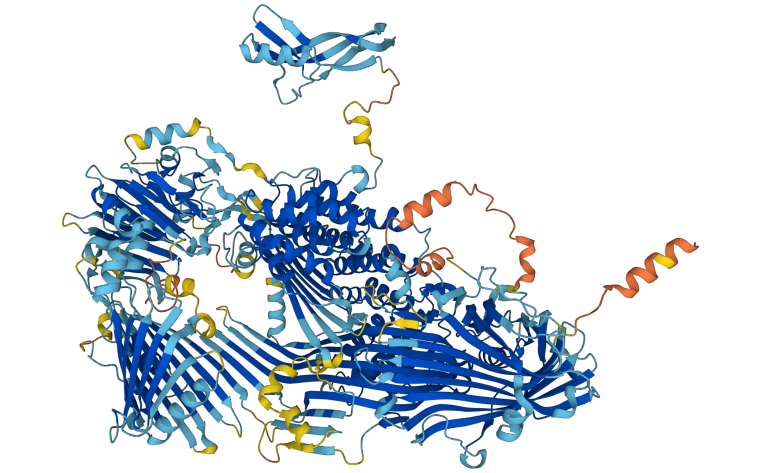
An artificial intelligence group says its program has successfully predicted the structure of nearly every protein known to science — effectively solving one of the "grand challenges" of biology and paving the way for new discoveries and technologies in fields as diverse as medicine, food security and climate science.
DeepMind, an AI firm owned by Google's parent company, Alphabet, announced Thursday that its AlphaFold program has expanded its open online database to include more than 200 million protein structures.
The vast catalog now encompasses the "entire protein universe," DeepMind CEO Demis Hassabis said in a news briefing — from the sequenced genomes of almost every organism on the planet.
Proteins are long, complex chains of amino acids that make up the building blocks of life. Scientists have long sought to untangle how these chains are elegantly twisted and folded into 3D shapes because understanding their structure can yield valuable insights into their function. Knowing a protein's specific shape and how its various molecules interact can, for instance, help researchers narrow down potential targets for medical treatments.
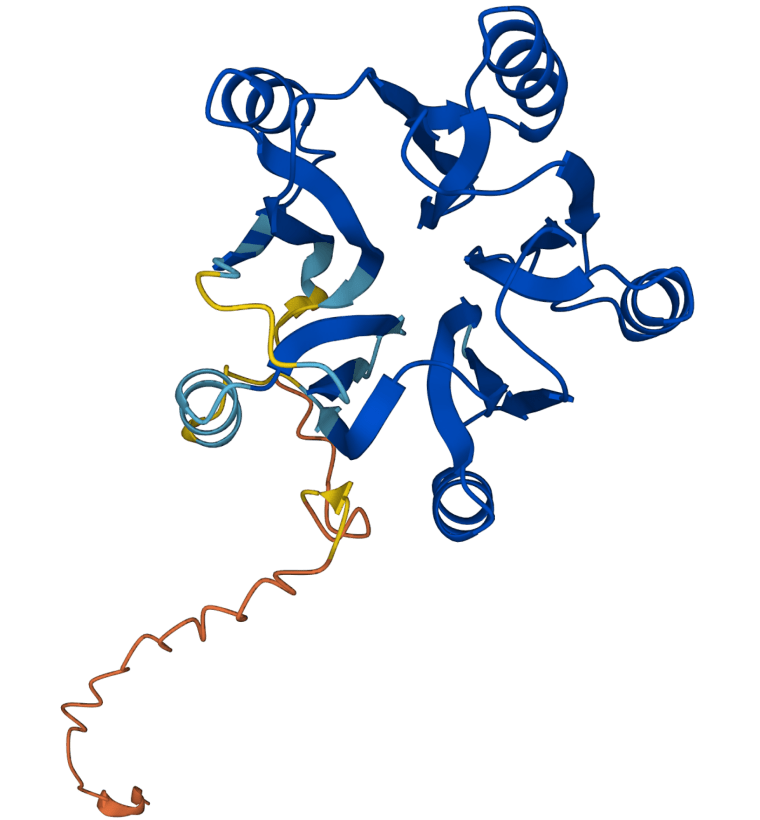
AlphaFold's upgraded database includes protein structures for plants, bacteria, animals and other organisms, according to DeepMind.
These updates offer "new opportunities for researchers to use AlphaFold to advance their work on important issues, including sustainability, food insecurity, and neglected diseases," Hassabis wrote in a blog post published Thursday about the milestone.
"By demonstrating that AI could accurately predict the shape of a protein down to atomic accuracy, at scale and in minutes, AlphaFold not only provided a solution to a 50-year grand challenge, it also became the first big proof point of our founding thesis: that artificial intelligence can dramatically accelerate scientific discovery, and in turn advance humanity," he wrote.
AlphaFold was introduced in 2020, and last year DeepMind wowed the scientific community by unveiling a catalog of structures that included virtually every protein in the human body. The so-called AlphaFold Protein Structure Database, built in collaboration with the European Molecular Biology Laboratory, included hundreds of thousands of newly predicted protein structures.
The rich trove of information is already being used by researchers around the world to study topics ranging from antibiotic resistance to plastic pollution, according to Hassabis.
Researchers at the University of Portsmouth in the United Kingdom, for example, announced in July 2021 that they are using the database to help engineer enzymes for recycling certain types of single-use plastics.
"AlphaFold provides us with an exciting new library of templates to engineer faster, more stable and cheaper enzymes for plastic recycling," John McGeehan, director of the University of Portsmouth's Centre for Enzyme Innovation, said in a statement at the time.
Hassabis said DeepMind is working to further expand its database, with particular emphasis on applications related to drug development, fundamental biology research, climate science, quantum chemistry and fusion.
"AlphaFold is a glimpse of the future," he wrote, "and what might be possible with computational and AI methods applied to biology."