New imaging method reveals whether antibiotics reach bacteria hiding in tissues
Researchers at the Francis Crick Institute and the University of Western Australia have developed a new imaging method to see where antibiotics have reached bacteria within tissues. The method could be used to help develop more effective antibiotic treatments, reducing the risk of antibiotic resistance.
During bacterial infections like tuberculosis, bacteria enter human cells, which poses a challenge for treatment, as antibiotics must reach and enter all infected cells in order to be effective. If researchers could select for or develop more effective antibiotics based on where they reach, this may reduce the length of treatment needed, which in turn could reduce the risk of antibiotic resistance developing.
In their study, published in PLoS Biology the researchers developed a new imaging method to see where in infected tissues and in cells an antibiotic given to treat tuberculosis reaches the bacteria. The scientists are continuing to work on the method, adapting it for other types of antibiotic and to image multiple antibiotics at the same time.
Max Gutierrez, author and group leader of the Host-Pathogen Interactions in Tuberculosis Laboratory at the Crick, says: "In the case of tuberculosis, people need to be treated with at least three different antibiotics over six months. We don't yet fully understand why this extended treatment is needed. We hope that being able to more clearly see where antibiotics are going, will help us better understand this process and find ways to improve it."
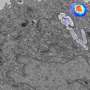
To develop the imaging method, called CLEIMiT, the researcher analyzed lung tissue from mice infected with tuberculosis and treated with the antibiotic bedaquiline.
They combined a variety of imaging methods, including confocal laser scanning microscopy, 3-D fluorescence microscopy, electron microscopy and nanoscale secondary ion mass spectrometry, to develop their new approach.
Using this method, they found that bedaquiline had not reached all infected cells in the lung tissue and also had not entered all infected areas within infected cells.
They also found this antibiotic was collecting in macrophages and in polymorphonuclear cells, both types of immune cell. This was a surprise as these cells have different environments and it wasn't thought that one antibiotic would be able to enter both.
Tony Fearns, author and senior laboratory research scientist in the Host-Pathogen Interactions in Tuberculosis Laboratory at the Crick, says: "Our approach could be used to help develop new antibiotics or to re-assess current antibiotics to judge how effectively they reach their targets. The more we learn about how drugs behave in the body, for example where they collect, the better we will be able to treat bacterial diseases like tuberculosis."
More information: Antony Fearns et al. Correlative light electron ion microscopy reveals in vivo localisation of bedaquiline in Mycobacterium tuberculosis–infected lungs, PLOS Biology (2020). DOI: 10.1371/journal.pbio.3000879
Journal information: PLoS Biology
Provided by The Francis Crick Institute