Abstract
The introduction of the CTLA-4 recombinant fusion protein has demonstrated therapeutic effects by selectively modulating T-cell activation in rheumatoid arthritis. Here we show, using a forward genetic approach, that a mutation in the SH3gl1 gene encoding the endocytic protein Endophilin A2 is associated with the development of arthritis in rodents. Defective expression of SH3gl1 affects T cell effector functions and alters the activation threshold of autoreactive T cells, thereby leading to complete protection from chronic autoimmune inflammatory disease in both mice and rats. We further show that SH3GL1 regulates human T cell signaling and T cell receptor internalization, and its expression is upregulated in rheumatoid arthritis patients. Collectively our data identify SH3GL1 as a key regulator of T cell activation, and as a potential target for treatment of autoimmune diseases.
Similar content being viewed by others
Introduction
Aberrant activation of autoreactive T cells has been suggested to initiate and drive autoimmune diseases, such as rheumatoid arthritis (RA) and multiple sclerosis (MS). It is believed that RA is initiated years before the clinical onset through an activation of major histocompatibility complex (MHC) class II-restricted autoreactive T cells that give specific help to B cells, leading to production of autoantibodies1. In RA, the chronic inflammatory attack of the joints, initiated by the adaptive immune system, is followed by an increased production of inflammatory cytokines such as TNF and IL-6 and during the last decades effective pharmaceutical treatments neutralizing this late inflammatory phase have been developed. Although early treatment has been shown to be more effective, as it improves the possibility to interfere with the underlying cause of autoimmune diseases, patients receive treatment well after disease is established2. Thus, a better understanding of the role of the adaptive immune system that drives the early stages of the disease i.e. before the disease is clinically overt, is needed to improve treatment of autoimmune diseases.
Genetic predisposition of RA is known to be strongly associated with MHC class II genes implicating a major contribution of T cells. Additionally, a recent genome wide meta-analysis revealed over 100 non-MHC risk loci where many were suggested to be related to T cell immune functions3. T cells can be found in abundance in the synovium of RA patients4 and the successful use of T cell targeting therapy, such as the CTLA-4-Ig fusion antibody abatacept, confirm the importance of T cells in the chronic stage of the disease. These observations are corroborated in animal models for RA, where depletion of T cells by use of antibodies or gene deletion protects against disease induction5,6. A specific role of the T cell receptor (TCR) in regulating development of arthritis has been further illustrated in the SKG mouse7, where a mutation in TCR signaling molecule Zap70 leads to altered TCR signaling. This in turn skews T cell development in the thymus increasing the number of autoreactive T cells leading to subsequent development of arthritis.
Herein, we have identified Endophilin A2 (EA2), encoded by the SH3gl1 gene, as a regulator of TCR internalization, signaling and downstream T cell effector functions. So far, EA2 has predominantly been studied in synaptic transmission in the central nervous system and in cancer8,9,10. We demonstrate that deficient expression of SH3gl1, caused by either a spontaneous mutation in the DA rat, or in genetically modified mice, leads to protection against autoimmunity. We hereby report that EA2 has a fundamental role in autoimmunity and limits the induction of autoreactive T cells. The discovery of the EA2’s impact on T cell activation opens up to explore new pathways and treatment possibilities for not only RA but for all T cell dependent inflammatory diseases.
Results
A spontaneous mutation in the SH3g11 gene protects against autoimmune arthritis
The DA rat is commonly used in autoimmunity research due to its high susceptibility to a number of chronic inflammatory disorders such as arthritis and experimental autoimmune encephalomyelitis11. We noticed an increased variability in susceptibility to arthritis in our inbred DA rat colony. Arthritis-resistant rats were selected and bred to establish a new line (denoted DA mutated (DAMut)), which was found to be completely protected against pristane-induced arthritis (PIA; Fig. 1a, b). We suspected the underlying cause to be of a genetic origin and not environmental. To test this hypothesis, the DAMut colony was re-established by cesarean sectioning into a SPF (FELASA II)- controlled facility. We injected the rats with pristane and followed arthritis development. The resulting clinical scores were similar to the conventional facility ruling out obvious environmental factors (Supplementary Fig. 1a, b). To genetically position the mutation, we crossed DAMut rats to the genetically different, arthritis susceptible, E3.DA-Pia457 rat strain12 and F2 offspring from this cross were immunized to induce PIA. Subsequent linkage analysis disclosed a significant association with arthritis incidence for a polymorphic marker on chromosome 9. Typing with additional markers lead to the identification of a quantitative trait locus (Pia43), at the telomeric end of chromosome 9 (Fig. 1c, d). The Pia43 locus had not been identified in previous E3xDA crosses13,14, confirming that this locus was unique to the DAMut line. The fragment was introgressed into a congenic strain DAMut.E3-Pia43 and minimized to 2 Mb by new recombinations (Fig. 1e, f).
a Mean arthritis score after pristane-induced arthritis in 5 DA and 7 DAMut rats. The arthritis data has been reproduced three times with the same results. b Incidence of arthritis after pristane-induced arthritis in DA and DAMut rats. c LOD score plot for incidence of PIA in 51 (E3.DA-Pia457x DAMut) x DAMut rats of all genotyped chromosomes. d LOD score plot for incidence of PIA in (E3.DA-Pia457 × DAMut) DAMut rats on chromosome 9, broken line at 2.63 indicate significant linkage e Schematic figure of the DAMut.E3-Pia43 congenic fragment on chromosome 9 with location of the SH3gl1 gene. Microsatellite markers in the Acsbg2 and Uxs1 genes indicate inner border of congenic fragment and markers Mllt1 and Dazl indicate outer borders. f Mean arthritis score after pristane-induced arthritis in 8 DAMut and 4 DAMut E3-Pia43 heterozygote littermates. Arthritis has been reproduced in several different congenic fragments with the same results. Non-parametrical Mann–Whitney U test was used for statistical evaluation of data. Data are presented as mean with error bars indicating ± SEM with each dot representing an individual value.
To identify the specific genetic alteration, we sequenced the DAMut rat genome and aligned it to the BN rat genome reference (Rno5). All variants in the DAMut.E3-Pia43 congenic region were manually compared to two previously sequenced DA genomes15,16. No single nucleotide polymorphism (SNP) or short insertion nor deletion (indel) was detected that could distinguish DAMut from the other DA strains in the congenic region, e.g., all 337 SNPs detected between DAMut and the reference sequence were also found in the other DA genomes. However, a structural variant was revealed, which was unique to the DAMut rat, and appeared to be the result of an insertion of a long terminal repeat (LTR) element of the ERV class I (ERV1) in intron 1 of the SH3gl1 gene (Fig. 2a).
a Schematic picture of the SH3gl1 gene with the location of the inserted ERVI long-term repeat element and location of the primer pairs used for quantification of the H3K4me3 and H4Ac levels. b Fold change of H3K4me3 levels in 3 DA and 3 DAMut naïve aged matched rats before (Primer pair 1 and 2) and after (Primer pair 3) the insertion in the SH3gl1 gene. c Fold change of H4Ac levels in 3 DA and 3 DAMut naïve aged matched rats before (Primer pair 1 and 2) and after (Primer pair 3) the insertion in the SH3gl1 gene. d Fold change of the SH3gl1 gene expression in PBMCs from 5 DA and 6 DAMut naïve aged matched rats. Relative fold change calculated to one DAMut reference sample. e Western blot analysis of the expression of the Endophilin A2 protein in brain samples of three DA and DAMut rats. Histone 2B was used as loading control. f Mean arthritis score after collagen-induced arthritis in 9 SH3gl1 knockouts and 16 wild-type littermates. Arthritis data has been reproduced twice with the same results. g Mean EAE score after spinal cord homogenate induced EAE in 10 DA and 10 DAMut rats. h Incidence of collagen-antibody induced arthritis in 10 SH3gl1−/− and 10 SH3gl1+/+ wild-type littermates. i Mean arthritis score of sick mice from above experiment after collagen-antibody induced arthritis in 4 SH3gl1 knockouts and three wild-type littermates. Non-parametrical Mann–Whitney U test was used for statistical evaluation of data. Data are presented as mean with error bars indicating ±SEM with each dot representing an individual value.
Retrotransposons such as LTR elements have been shown to regulate gene expression in both mice and man17. To investigate if this was also the case for the ERV1 insertion in the SH3gl1 gene in DAMut rats, we determined the levels of lysine 4 methylation in histone 3 (H3K4me3) and the acetylation of histone 4 (H4Ac) using chromatin-immunoprecipitation (ChIP) and qPCR, as these modifications have been associated with active gene transcription18,19. We observed that H3K4me3 and H4ac levels upstream of the inserted LTR were similar in DA and DAMut rats. In contrast, levels in regions situated downstream of the insertion were down-regulated (Fig. 2B, C), indicating that SH3gl1 gene transcription was not active in DAMut rats. To determine if this had any impact on gene expression we analysed peripheral blood monocytic cells (PBMCs) from DA and DAMut rats and quantified gene expression with qPCR. Strikingly, the DAMut rats had almost no expression of the SH3gl1 gene compared to DA rats (Fig. 2d). This vastly reduced gene expression also translated to a reduced level of the SH3gl1 encoded protein EA2 as determined by western blotting (Fig. 2e). To confirm that the arthritis resistance observed in the DAMut rats was due to a deficiency in EA2 expression we introgressed a deletion of the SH3gl1 gene into arthritis-susceptible B6N.Q mice through backcrossing and evaluated them with collagen-induced arthritis (CIA). Similar to the DAMut rats, the SH3gl1 deficient mice were also protected from arthritis, in contrast to their SH3gl1 sufficient wild-type littermates (Fig. 2f). To investigate whether the effect was restricted to control only arthritis development or also other autoimmune diseases, we tested experimental autoimmune encephalomyelitis (EAE), a T-cell-dependent model of MS, which confirmed the EA2 mediated protective effect seen in arthritis (Fig. 2g). Additionally, we tested the T-cell independent collagen-antibody induced arthritis model20 and observed no difference in disease severity or induction (Fig. 2h, i). We conclude that EA2 is a major regulator of T cell dependent autoimmune disease.
EA2 deficiency alters the induction threshold of autoreactive T cells
Since EA2 deficiency had such a great impact on T cell dependent autoimmune disorders we investigated if the expression of SH3gl1 increased in T cells after arthritis induction. Indeed 8 days after in vivo activation the expression of SH3gl1 had more than doubled (Fig. 3a) suggesting an important function of SH3gl1 during T cell priming and activation. To further elucidate if the protection in SH3gl1 deficient rats is mediated by T cells we used the CD4+ αβ+ T cell dependent pristane-induced adoptive transfer model21. Pristane-primed lymph node cells from DA or DAMut rats were expanded for T cells ex vivo before transfer into naïve DA recipients. Only cells from DA rat donors induced severe arthritis (Fig. 3b), indicating that the DAMut rats were not able to generate arthritogenic T cells. To confirm that the arthritis resistance was intrinsic to T cells, we transferred thymocytes from SH3gl1 deficient or wild-type littermate mice into TCRβ knockout mice before the induction of glucose-6-phosphate isomerase (GPI) induced arthritis. Mice receiving SH3gl1 deficient thymocytes developed nearly no arthritis (Fig. 3c) and had fewer antigen-specific cells compared to the wild-type recipients as determined by an in vitro recall assay using the GPI peptide while no significant difference could be seen in mitogen activated cells (Fig. 3d). To determine if this reduction in antigen-specific cells was due to a defect in the thymic T cell development, we assessed thymic T cell populations by flow cytometry. No difference in T cell populations could be observed between the SH3gl1 deficient mice compared to their wild-type littermates indicating that the reduced numbers of antigen-specific cells were not due to a defect in thymic T cell development (Fig. 3e) but were instead induced in the periphery. To better understand why the SH3gl1 deficient T cells do not become arthritogenic we looked at activation markers and induction of anergy. While no significant differences in activation markers could be seen ten days after GPI-peptide immunization, the SH3gl1 deficient T cells significantly upregulated anergy markers like PD-1 and CD73/FR4 (Fig. 3f). In a subsequent experiment comparing responses to GPI protein and ovalbumin, the same trend was observed for GPI but not for the exogenous antigen ovalbumin (Supplementary Fig. 2). We next investigated if the SH3gl1 deficient T cells had a regulatory or suppressive function in the pristane-induced T cell transfer model. Naïve DA rats were given either pristane-primed DA cells alone or together with an equal number of DAMut cells, with DAMut cells alone serving as a negative control. No reduction in arthritis severity was evident in recipients receiving pristane- primed DA cells together with DAMut cells, indicating that DAMut T cells do not exert their regulatory function via active suppression (Fig. 3g, h) but rather via inhibited activation.
a Expression of SH3gl1 in sorted T cells from lymph nodes from DA rats at Day 0 and Day 8 after pristane immunization. b Mean arthritis score after adoptive transfer of pristane-primed DA and DAMut cells to 9 respectively 4 naïve DA recipients. The adoptive transfer experiment has been reproduced a total of three times with the same results. c Mean arthritis score after GPI-induced arthritis in 17 and 19 TCRβ knockout recipients that had received either SH3gl1−/− or SH3gl1+/+ thymocytes respectively, 7 days prior to disease induction. Data is pooled from three individual experiments. d Number of antigen specific IL-2, IL-17A and IFNg producing T cells after recall stimulation with GPI peptide and IL-2 producing cells after ConA stimulation day 44 after GPI-induced arthritis. Data from 9 TCRβ knockout mice reconstituted with wild-type T cells and 8 TCRβ knockout mice reconstituted SH3gl1 T cells. Recall experiment has been repeated twice in SH3gl1−/− mice. e Frequency of double negative (DN), double positive (DP), single positive CD4 (SP CD4), and single positive CD8 (SP CD8) thymocytes in 8 SH3gl1−/− and 8 SH3gl1+/+ mice. Data has been reproduced twice. f Frequency of CD25, CD44, PD-1, CD73+FR4+, Ki-67, and FoxP3 positive cells in CD4+TCRb+ cells from inguinal lymph nodes ten days after GPI-peptide immunization from 6 SH3gl1−/− and 6 SH3gl1+/+ mice. g Mean arthritis score in naïve DA recipients after adoptive transfer of pristane-primed DAMut cells only, DA cells only or DA cells in addition with DAMut cells, 7 rats per group. h Arthritis sum score of the DA recipients above after adoptive transfer of pristane-primed DA cells only or addition with DAMut cells, 7 rats per group. Non-parametrical Mann–Whitney U test was used for statistical evaluation of data. Data are presented as mean with error bars indicating ±SEM with each dot representing an individual value.
EA2 deficient rodents develop normally and mount a normal response to cancer and infection
EA2 is a ubiquitous protein with high expression in the central nervous system22. Thus, EA2 deficiency could potentially affect many cell types and impact development. However, we did not observe any major physiological disturbances caused by EA2 deficiency in DAMut rats by general measurements such as blood cell counts, body weight, breeding capacity and life span (up to 1.5 years) in which the DAMut rats were identical to their normal DA littermates (Supplementary Fig. 3a–e). Similarly to the DAMut rats, the SH3gl1 deficient mice develop normally as compared to their wild-type littermates8. Given that the EA2 deficient rodents exhibited a profound protection against autoimmune diseases, we assumed that they could have a severely defective immune system. Our animal facility is specific pathogen free but had an outbreak of S. aureus caused infections which provoked septic arthritis in immunodeficient mice23. However, no SH3gl1 deficient animals developed a clinical S. aureus infection during this outbreak. To further challenge the SH3gl1 deficient mice towards bacterial infections, an arthritogenic S. aureus LS-1 strain24 was inoculated intravenously. This septic arthritis model using S. aureus LS-1 strain has been shown to be dependent on T cells via activation of T cells by the superantigen, toxic shock syndrome toxin-1 (TSST-1)25. In this model of septic arthritis, we did not observe any difference in bacterial clearance or development of arthritis in SH3gl1 deficient mice compared to wild-type littermates (Supplementary Fig. 3f, g). To further challenge the immune system in EA2 deficient mice, we injected SH3gl1 deficient and wild-type littermate mice with B16F10 melanoma cells and followed tumor growth. No excessive tumor growth was observed in EA2 deficient mice compared to wild-type littermates (Supplementary Fig. 3h). Thus, EA2 deficiency seems to affect regulation of autoimmunity but does not affect the general health of the rodents nor their susceptibility to a bacterial infection caused by S. aureus nor an excess growth of cancer cells.
EA2 deficiency leads to reduced T cell activation via the TCR
Considering the endocytic function of EA2 and the profound effect that loss of EA2 expression had on T cells specifically, we reasoned that EA2 might be important for the internalization of the TCR. Thus, we checked whether EA2 co-localized with the TCR upon activation using a proximity-ligase assay. While no co-localization could be found in un-stimulated cells, we could see that EA2 and the TCR co-localized after only three minutes of anti-CD3/CD28 stimulation (Fig. 4a and Supplementary Fig. 4). We next studied the kinetics of the TCR internalization and indeed, SH3gl1 deficient T cells internalized their TCRs at a slower rate compared to SH3gl1 sufficient T cells (Fig. 4b) with a reduced internalization seen already at 15 min after stimulation. The observed decreased rate of TCR internalization was not due to an increased recycling of TCR as differences in TCR internalization between the SH3gl1 deficient and sufficient T cells sustain even after Brefeldin A treatment (Fig. 4c). To investigate if this had any impact on the responsiveness of the T cells we investigated downstream TCR signaling molecules in SH3gl1 deficient and wild-type T cells. A reduced TCR signaling cascade was observed in SH3gl1 deficient T cells compared to wild-type T cells, with reduced levels of phosphorylated Zap70 and activation of ERK1/2 (Fig. 4d–f). This reduction in responsiveness in the SH3gl1 deficient T cells translated to reduced T cell proliferation following in vitro stimulation via CD3/CD28 (Fig. 4g, h). The observed reduced proliferation was not the result of reduced internalization of the IL-2 receptor as previously described26 but must stem from a reduced activation via the TCR (Supplementary Fig. 5).
a EA2 co-localization with the TCR in Jurkat cells after anti-CD3/CD28 stimulation determined by proximity-ligase assay and visualized as TexasRed+ spots using confocal imaging. b Percentage of internalized TCR in T cells from 5 DA and 5 DAMut rats after anti-CD3/CD28 stimulation. Data has been reproduced two times. c Percentage of internalized TCR in CD4+CD3+ T cells from three SH3gl1−/− and SH3gl1+/+ mice stimulated with anti-CD3/CD28 for 30 min in the presence or absence of Brefeldin A at 1.25 μg/ml. Experiment has been repeated twice with the same results. d Representative Western blot analysis from two experiments of phosphorylated and unphosphorylated ZAP70 and ERK1/2 in T cells from two SH3gl1−/− and SH3gl1+/+ mice stimulated with anti-CD3/CD28 for 0, 3, and 7 min. Histone 2B was used as loading control. e Normalized OD values of phosphorylated Zap70 compared to unphosphorylated Zap70 from two SH3gl1−/− and SH3gl1+/+ mice. f Normalized OD values of phosphorylated ERK1/2 compared to unphosphorylated ERK1/2 from two SH3gl1−/− and SH3gl1+/+ mice. g Flow cytometry blot showing in vitro cell proliferation of DAMut and DA T cells 72 h after anti-CD3/CD28 stimulation. h Division index of sorted T cells from 7 DA and 5 DAMut rats after 72 h of anti-CD3/CD28 stimulation. Non-parametrical Mann–Whitney U test was used for statistical evaluation of data. Data are presented as mean with error bars indicating ± SEM with each dot representing an individual value.
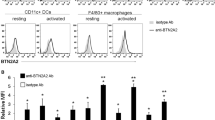
SH3GL1 regulates TCR responses in human T cells and its expression is upregulated in RA patients
Since EA2 is highly conserved and had such a profound effect on T-cell-mediated arthritogenicity in both mice and rats, we next investigated to what extent SH3GL1 is of importance in human T cells. To demonstrate the relevance of EA2 in human T cells we made SH3GL1 CRISPR knock-out Jurkat T cells (Supplementary Fig. 6) and stimulated them in vitro with anti-CD3/CD28. Both TCR signaling and TCR internalization were reduced in SH3GL1 deficient Jurkat cells compared to wild-type control and normal Jurkat T cells (Fig. 5a, b). We addressed the possibility that SH3GL1 could be overexpressed in T cells from RA patients and determined the expression of both SH3GL1 and the TCR molecule CD3e from whole blood of RA patients and healthy controls and found that the SH3GL1 expression correlated with expression of the CD3E molecule (Fig. 5c). Similar to the findings in animal models, the levels of SH3GL1 was higher in RA patients when normalized for CD3E (Fig. 5d) corroborating the data from the NCBI GEO database27 where SH3gl1 gene expression was found to be upregulated in sorted CD4 + T cells from RA patients compared to healthy control (Fig. 5e). Thus, SH3GL1 has a conserved mechanistic function in T cells across species and is relevant in a human disease setting.
a TCR internalization in SH3GL1 CRISPR knockout Jurkat cells (T3-5 and T3-21) and wild-type control (T3-23). b Representative western blot analysis from two experiments of phosphorylated ZAP70 and ERK1/2 in T cells from in SH3GL1 CRISPR knockout (T3-5 and T3-21), wild-type control (T3-23), and Jurkat (J) cells with anti-CD3/CD28 for 0, 3, and 7 min. Histone 2B was used as loading control. c Correlation of CD3e and SH3GL1 gene expression in whole blood of healthy controls and RA patients. Each circle represents one individual. d Normalized SH3GL1 gene expression to CD3e gene expression in whole blood of 35 healthy controls and 36 RA patients, randomly selected from the cohort used in c. Each circle represents one individual. e SH3GL1 gene expression in sorted CD4+ T cells from 10 healthy controls and 8 RA patients (data accessible at NCBI GEO database, accession GSE4588). Non-parametrical Mann–Whitney U test was used for statistical evaluation of data. Data are presented as mean with error bars indicating ±SEM with each dot representing an individual value.
Discussion
Aberrant activation of autoreactive T cells is the key for the self-perpetuating vicious circle of activation of both the innate and adaptive immune response that leads to chronicity and lack of resolution in autoimmune diseases. Using a rat animal model of RA, we discovered SH3GL1 as a major regulator of T cell effector function and autoimmune diseases. We show that deficiency of SH3gl1 leads to a complete protection against autoimmune diseases in both mutated DA rats and in SH3gl1 knockout mice and that this results from loss of T cell effector functions. Furthermore, we show that SH3gl1 expression was increased in T cells during autoimmune arthritis in both rodents and RA patients.
TCR internalization and surface recycling following peptide-MHC recognition on the antigen presenting cell plays a pivotal role in establishing a stable immunological synapse and subsequent T cell activation28. To limit potential induction of autoreactive responses, T cell activation is tightly regulated via the TCR and co-stimulatory molecules. Both the surface expression of the TCR as well as the adapters in the TCR signaling cascade are of importance in dictating the effector functions of the T cells. For example, CD4+ T cells deficient in the WASH protein show reduced trafficking of the TCR and proliferation of T cells. Similar to SH3gl1 deficient animals, CD4+ conditional WASH knockout mice are also protected from EAE29. Several earlier studies demonstrate that SH3GL1 is necessary for internalization and trafficking of a number of receptors26,30,31. Our data demonstrate that SH3GL1 also regulates the internalization of the TCR. Because of the importance of TCR internalization and formation of the immunological synapse for efficient TCR signaling, a lack of SH3GL1 leads to reduced T cell signaling and would subsequently result in reduced numbers of autoreactive T cells, in turn limiting the arthritis development. The importance of adapters in the TCR signaling cascade was recently demonstrated by targeting the adapter protein NCK32. Like in SH3gl1 deficient animals, inhibition of NCK reduce proliferation of the T cells and leads to induction of immunosuppression and protection against EAE. Although the regulation of the TCR is different between SH3GL1 and NCK the resulting outcome seems to be similar. Development of effective tissue restricted immunotherapies in the field of autoimmune diseases has proven difficult, with a variety of antigen-specific vaccination strategies developed in the 1990s with limited success33,34. Consequently, instead of attempting to address antigen specificity, the field moved more into the depletion of molecules (e.g. TNF blockade35) or whole cell populations (e.g. B cells using anti-CD20 antibody36) involved in the pathogenesis. Although these therapies have been beneficial they are not curative and there is still a substantial number of patients who are non-responders or develop severe side-effects such as increased susceptibility to infections37,38. Our results with SH3gl1 in rodents demonstrate an interesting pathway by which activation of autoreactive T cells is modulated via the TCR while retaining the ability to respond to pathogens. Thus, SH3GL1 targeting could be a promising therapeutic treatment strategy for RA as well as in other T cell-mediated diseases.
Methods
Animals
The E3/ZtmRhd and DA/ZtmRhd rats originated from the Zentralinstitute für Versuchstierzucht, Hannover, Germany and were bred for more than 20 brother/sister mating generations in the animal facility of Medical Inflammation Research lab. The DAMut rats were isolated from the DA/ZtmRhd stock and bred for more than 10 brother/sister mating generations. To obtain the (E3.DA-Pia457x DAMut) DAMut rats used in the linkage analysis, 2 male DAMut rats were bred with 4 female E3.DA-Pia45712 rats. Subsequently, 10 female F1 hybrids were bred with 6 DAMut male rats to produce 51 males of the F2 offspring. The C57BL/6.129-Sh3gl1tm1Pdc/J8, titled SH3gl1−/− in paper, mice were a kind gift from Professor Pietro de Camilli and backcrossed for five generations to the arthritis susceptible C57BL/6 mouse, expressing MHC class II A(q)39. C57BL/6.129P2-Tcrbtm1Mom/J (Stock No: 002118), titled TCRb−/− in paper, mice were bought from Jackson Laboratory and backcrossed to C57BL/10 expressing the MHC class II A(q) for more than ten generations. All animals included in the experiments were kept in a specific pathogen-free environment following the Federation of European Animal Laboratory Science Association guidelines (FELASA II), in a climate-controlled environment with a 12-h light/dark cycle and fed standard rodent chow and water ad libitum. All animal experiments followed the ARRIVE guidelines, they were performed blindly, age- and sex balanced, mixed in cages and with littermate controls, and approved by the local ethical committees (Malmö/Lund, Göteborg and Stockholm, Sweden, ethical permits M109/07, M107/07, N67/10, N69/10, N169/10, N490/12, N134/13, 353-2012, N35/16, and N288/15).
Patients and healthy controls
Whole blood from RA patients and healthy controls matched for gender, age and ethnicity were collected in PAX tubes at the Rheumatology Clinic at Karolinska University hospital. RA patients all met the 1987 American college of rheumatology criteria for diagnosing RA. Informed consent was obtained from all the participants and the Stockholm ethical review board approved the study.
Experimentally induced arthritis
Pristane-induced arthritis (PIA) was induced by a single injection of 100 μl pristane (2, 6, 10, 14-tetramethylpentadecane; ACROS Organics) at the base of the tail. Collagen-induced arthritis (CIA) was induced by injection of 100 μg of pepsin-digested rat collagen type II (as described in40) emulsified in 100 μl complete Freund’s adjuvant (Difco) with a boost injection at day 35 with collagen type 2 in incomplete Freund’s adjuvant (Difco). Glucose-6-phosphate isomerase (GPI)-induced arthritis was induced by a single injection of synthetically produced peptide corresponding to the human GPI protein aa325-339(hGPI325-339) emulsified in complete Freund’s adjuvant (Difco)41. S. aureus-induced arthritis was induced by inoculation of the S. aureus TSST-1-producing; LS-1 strain intravenously in one of the tail veins with 2.07 × 107 S. aureus LS-1/ml in a total volume of 200 μl phosphate-buffered saline (PBS). Arthritis development in PIA, CIA, and GPI experiments were monitored using a macroscopic scoring system. Each limb with the highest possible count of 15, thus with a total possible score of 60 per animal described in more detail for rats in42 and for mice in43. The S. aureus-induced arthritis development was monitored using a macroscopic scoring system where each limb was scored according to a scoring scheme (0, neither swelling nor erythema; 1, mild swelling and/or erythema; 2, moderate swelling and erythema; and 3, marked swelling and erythema). The total score was calculated by adding up all the scores within each animal tested.
Experimental autoimmune encephalomyelitis
Spinal cord from naïve DA rats was taken and homogenized. EAE was induced by a single injection of spinal cord homogenate emulsified in incomplete Freund’s adjuvant (Difco). EAE was macroscopically scored according to the following scoring scheme 0 = Normal, 1 = Tail weakness, 2 = Tail paralysis, normal gait, 2.5 = Tail paralysis, little affected gait, 3 = Tail paralysis, low back and mild waddle, 3 .5 = Tail paralysis and low back, severe waddle, 4 = Tail paralysis, severe waddle, less sure footing, 4.5 = Tail paralysis, severe waddle, falling and lost balance, 5 = Tail paralysis and paralysis of one limb, crawling, 6 = Tail paralysis and paralysis of a pair of limbs, back is affected, 7 = Tetraparesis, and 8 = Pre-morbid or deceased.
Adoptive T cell transfer of arthritogenic cells
Eight days after pristane injection, rats were euthanized by CO2 inhalation and inguinal lymph nodes were taken and mechanically homogenized through 40μm filters. Cells were washed in PBS and T cells reactivated and expanded in vitro in D-MEM supplemented with HEPES (GIBCO), streptomycin/D-penicillin (104 IU/ml penicillin, 10 mg/ml streptomycin; Invitrogen Life Technologies), β-mercaptoethanol (GIBCO), 5% fetal calf serum (GIBCO) and Concavalin A (3 µg/ml; Sigma-Aldrich). Cells were incubated at 37 °C and 5% CO2 for 48 h. Cells were washed in PBS and transferred to naïve DA rats.
Genotyping and linkage analysis
Toe biopsies were sampled and DNA was prepared by alkaline lysis. The DNA was amplified using fluorescence-marked microsatellites and standard polymerase chain reaction (PCR) reagents (dNTPs, MgCl2, Taq Polymerase). The solution was assayed in a PCR Thermal Cycler and the final PCR products were pooled and size-fractioned on a MegaBACE 1000 (Amersham Pharmacia Biotech, Uppsala, Sweden) alternatively on a ABI3730 DNA Analyzer (Applied Biosystems, Life Technologies Corporation, Carlsbad, USA). The data was analyzed using the enclosed program Genetic Profiler 1.1 respectively GeneMapper® Software v4.1 Sequences for microsatellite markers used in the linkage analysis were retrieved from Rat Genome Database (http://rgd.mcw.edu/rgdweb/search/markers.html?100). Additional primers used in mapping the congenic fragments were created using genomic sequences from publicly available rat sequences at NCBI Gene (http://www.ncbi.nlm.nih.gov/gene). Microsattellites were identified and primers were created using the primer design program, Primer3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi). Primer specificity was controlled by running the sequences through Primer-BLAST at NCBI. (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Oligos were later ordered from Eurofins MWG Operon. Linkage analysis was performed using R (The R Foundation of Statistical Computing, version 2.0.1) and R/qtl44. The normal model and Haley-Knott regression method (1-cM steps) were used for all calculations. Significance threshold of logarithm of the odds (LOD) was determined using permutation tests (n = 10000), in which p < 0.05 was considered significant. The genetic map was generated in the R/qtl environment based on the recombinations in the cross.
Chromatin-immunoprecipitation and quantitative PCR
Based on previous ChIP-Seq results from BN and SHR rats45, we identified regions with high levels of H3K4me3 modifications in close proximity to the Sh3gl1 gene. Spleen powder (100 mg) from 3 DA and 3 DAMut rats were used for nuclei isolation and subsequent ChIP analysis was performed according to45. The regions of interest were analyzed by ChIP followed by qPCR on ABI 7900HT detection system (Applied Biosystems). PCR primers were designed to amplify designated genomic regions using Primer Express software (Applied Biosystems). qPCR assays were carried out in 384-well plates with a final volume of 20 µL each for 40 cycles. We used Power SYBR Green PCR Master Mix (Applied Biosystems) with diluted ChIPed DNA or un-enriched input DNA as template. Enrichment ratios were calculated according to the 2-∆∆Ct method with endogenous controls (Gapdh) similar to gene expression data sets. For H3K4me3 and H4ac modification see supplementary Table 1 for sequences of primers used for quantification
RNA extraction and quantitative- polymerase chain reaction
Mononuclear leukocytes from blood (separated from Ficoll-density gradient) or sorted T cells (Pan T Cell MicroBeads, Miltenyi Biotec) were obtained from naïve DA and DAMut rats and RNA was isolated (Trizol and Pure-link mRNA kit, Invitrogen). Whole blood from RA patients and healthy controls were collected in PAXgene Blood RNA Tubes and total RNA was extracted with the PAXgene Blood RNA kit (PreAnalytiX, Feldbachstrasse, Switzerland) according to the manufacturer’s protocol. cDNA conversion was performed with an iScript cDNA synthesis (Bio-Rad, Hercules, CA, USA). Gene expression experiments were performed using TaqMan gene expression assays(Hs04235263_g1(Sh3gl1), Hs04194521_s1 (PPIA), Hs01062241_m1(CD3e), Rn01527769_g1(Sh3gl1), Rn01527840_m1(Hprt1), Applied Biosystems) and samples were run on a CFX96 RT-PCR(Bio-Rad) according to manufacturers description. Relative fold change was calculated according to the 2-∆∆Ct method after normalization to reference gene, Hprt1 for rat gene expression and PPIA for the human gene expression experiments.
Western blotting
Whole cell lysates from DA and DAMut rats were obtained from brain samples by lysis in RIPA buffer (ThermoScientific). Samples were run in a NuPAGE 4–12% Bis-Tris gel (Novex, Invitrogen) and transferred to a PVDF membrane (EMD, Millipore). Blots were blocked with 5% bovine serum albumin (Sigma-Aldrich) in Tris-buffered saline with 0.1% Tween 20 (Cell Signaling Technology) and subsequently incubated with mouse anti-rat Endophilin A2 (clone S51-1, Origene Technologies, Inc) and rabbit anti-histone 2B (Cat. No C49810, LifeSpan Biosciences, Inc). Blot was incubated with peroxidase-conjugated goat anti-mouse IgG(Cat no. 115-035-062, Jackson ImmunoResearch laboratories Inc.) or peroxidase-conjugated donkey anti-rabbit IgG (Cat no.771-036-152, Jackson ImmunoResearch laboratories Inc.) respectively and developed using enhanced chemiluminescence. Pictures including the entire blot can be found in the Source Data file.
Determining number of antigen-specific cells with enzyme-linked immunospot (ELISpot) assay
Single cell suspension from spleens of mice previously immunized for GPI arthritis was added to plates (MSIPS4W10, Millipore) previously coated with antibodies to IL-2 (clone JES6-IA12), IL-17(clone TC11-18H10) and a-IFNg(AN18) and stimulated for 48 h with 10 μM hGPI325-339 in D-MEM supplemented with HEPES (GIBCO), streptomycin/D-penicillin (104 IU/ml penicillin, 10 mg/ml streptomycin; Invitrogen Life Technologies), b-mercaptoethanol (GIBCO), 10% fetal calf serum (GIBCO). Plates were washed and incubated with biotinylated antibodies to IL-2 (clone 5H4), IL-17 (clone TC11-8H4) and IFNγ (R46A2). Plates were washed and Extravidin-Phosphatase (Sigma) was added to wells and incubated for 45 min and washed. Plates were subsequently developed using BCIP/NBT(Sigma). Plates were scanned, analyzed and number of reactive cells were counted with ImmunoSpot software (Cellular Technology Ltd.).
T cell development
Single cell suspension from thymus of C57BL/6.129-Sh3gl1tm1Pdc/J.Q and their wild-type littermates were stained with anti-mouse monoclonal antibodies (B220-Fitc, CD5-PE, CD25-PECy7, TCRb-PerCP5.5, CD4-Q605, CD8-BV650, CD69-APC, CD44-A750) for 20 min at 4 °C. After washing with PBS, cells were acquired on a LSRII (BD Biosciences, Franklin Lakes, NJ, USA) using the BD FACSDiva™ software (BD Biosciences) with gates set to include all viable cells and later analyzed by FlowJo (Tree Star, Inc.) software.
Cell proliferation assays
T cells from DA and DAMut lymph nodes were negatively sorted using biotinylated mouse anti-rat CD11b/c (Clone OX42, Biolegend) and mouse anti-rat CD45Ra (Clone OX33, Biolegend) and anti-biotin MicroBeads (Miltenyi Biotec). Sorted T cells were then labeled with CellTrace-Violet (Molecular Probes, Invitrogen) and added to plates coated with anti-rat αβTCR(Clone R73) and anti-rat CD28(Clone JJI319) in D-MEM supplemented with HEPES (GIBCO), streptomycin/D-penicillin (104 IU/ml penicillin, 10 mg/ml streptomycin; Invitrogen Life Technologies), b-mercaptoethanol (GIBCO), 10% fetal calf serum (GIBCO) and incubated at 37 °C, 5% CO2. After 72 h cells were washed and later acquired on a LSRII (BD Biosciences, Franklin Lakes, NJ, USA) using the BD FACSDiva™ software (BD Biosciences) with gates set to include all viable cells and later analyzed by FlowJo (Tree Star, Inc.) software.
TCR internalization assay
Lymph node cells from DA and DAMut rats were stained with mouse anti-rat CD3 (Clone 1F4) conjugated with Fitc (Biolegend) at 4 °C. Labeled cells were added to plates coated with anti-rat αβTCR (Clone R73) and anti-rat CD28(Clone JJI319) in D-MEM supplemented with HEPES (GIBCO), streptomycin/d-penicillin (104 IU/ml penicillin, 10 mg/ml streptomycin; Invitrogen Life Technologies), b-mercaptoethanol (GIBCO), 10% fetal calf serum (GIBCO), and incubated for 15, 30, and 60 min at 37 °C or left on ice for zero time point. Cells were washed and spilt into two fractions, were one fraction was stripped of surface anti-CD3-Alexa488 antibodies with PBS (GIBCO) pH 2 for 1 min and the other fraction left untreated. For the TCR internalization assay with the CRISPR Jurkat T cells the cells were stained with anti-human CD3 (Clone UTCH1) conjugated with Alexa488 (BD Biosciences) at 4 °C. Labeled cells were added to plates coated with anti-human CD3 (Clone Hit3a, BD Biosciences) and anti-human CD28(Clone CD28.2, BD Biosciences) in RPMI1640 (GIBCO), supplemented streptomycin/D-penicillin (104 IU/ml penicillin, 10 mg/ml streptomycin; Invitrogen Life Technologies) and 10% fetal calf serum (GIBCO) and incubated for 15, 30, and 60 min at 37 °C or left on ice for zero time point. Cells were washed and spilt into two fractions, were one fraction was quenched of surface anti-CD3-Alexa488 signal with unlabeled anti-Alexa488 (MolecularProbes, Invitrogen) and the other fraction left untreated. For the Brefeldin A inhibitor experiment lymph node cells from SH3gl1−/− and SH3gl1+/+ mice were stained with Armenian hamster anti-mouse TCRb (Clone H57-597, Biolegend) conjugated with Alexa488 at 4 °C. Labeled cells were added to plates coated with anti-mouse CD3e (Clone 145-2C11, BD Biosciences) and anti-mouse CD28(Clone 37.51, BD Biosciences) in D-MEM supplemented with HEPES (GIBCO), streptomycin/D-penicillin (104 IU/ml penicillin, 10 mg/ml streptomycin; Invitrogen Life Technologies), b-mercaptoethanol (GIBCO), 10% fetal calf serum (GIBCO) and together with 1, 25ug/ml Brefeldin A (Sigma) and incubated for 30 min at 37 °C or left on ice for zero time point. Cells were washed and spilt into two fractions, quenched or no quench, and subsequently stained with rat anti-mouse CD45R-PE-Cy7 (Clone RA3-6B2, BD Biosciences), rat anti-mouse CD4-BV605 (Clone RM4.5, BD Biosciences), rat anti-mouse CD3-PacificBlue (Clone 17A2, Biolegend) and anti-Alexa488 (MolecularProbes, Invitrogen) to half of the samples (quenched). Cells were acquired on a LSRII (BD Biosciences, Franklin Lakes, NJ, USA) using the BD FACSDiva™ software (BD Biosciences) with gates set to exclude doublets and include all viable cells determined as LIVE/DEAD™ -Near-IR (Invitrogen) negative cells and later analyzed by FlowJo (Tree Star, Inc.) software. Percentage of internalized CD3-Alexa488 was calculated as (Qx-Q0)/(Tt-Q0)x100, where Qx is the mean fluorescence of cells quenched with anti-Alexa488 at each time point, Q0 is the mean fluorescence of cells quenched at time zero, and Tt is the mean fluorescence of cells that were not quenched.
TCR signaling
T cells from lymph nodes and spleens of C57BL/6.129-Sh3gl1tm1Pdc/J.Q mice and their wild-type littermates were sorted (Untouched CD4 + T cells MicroBeads, Miltenyi Biotec) and resuspended in D-MEM supplemented with HEPES (GIBCO), streptomycin/D-penicillin (104 IU/ml penicillin, 10 mg/ml streptomycin; Invitrogen Life Technologies), b-mercaptoethanol (GIBCO), 10% fetal calf serum (GIBCO). Cells were rested for one hour at 37 °C and later added to plates coated with 10 μg/ml rat anti-mouse CD3(Clone 145-2C11, BD Biosciences) and 5 μg/ml rat anti-mouse CD28(Clone 37.51, BD Biosciences) and stimulated for 3 and 7 min. Cells were lysed using M-PER(ThermoScientific) supplemented with HALT phosphatase and protease inhibitors (ThermoScientific). Samples were run on a NuPAGE 4-12% Bis-Tris gel (Novex, Invitrogen) and transferred to PVDF membrane (EMD, Millipore). Blots were blocked with 5% bovine serum albumin (Sigma-Aldrich) in Tris-buffered saline with 0.1% Tween (Cell Signaling Technology) and subsequently incubated with rabbit anti- mouse phosphorylated Zap70 (Clone 65E4, Cell Signaling Technology), rabbit anti- phosphorylated-p44/42 MAPK (Erk1/2; Thr202/Tyr204; Cell Signaling Technology) and rabbit anti-histone 2B (Cat no. ab1790 Abcam) as loading control. Blot was incubated with peroxidase-conjugated donkey anti-rabbit IgG (Cat no.771-036-152, Jackson ImmunoResearch laboratories Inc.) and developed using enhanced chemiluminescence. Pictures including the entire blot can be found in the Source Data file.
Whole genome sequencing and bioinformatics analysis
Genomic DNA from liver from DAMut rats was isolated using the nuclear lysis buffer, followed by ethanol precipitation of the DNA. DNA integrity was validated with gel electrophoresis. Next generation sequencing was performed at Science for Life laboratory, Stockholm, Sweden using Illumina HiSeq 2000 with 650 bp insert standard DNA (Illumina TruSeq DNA) and paired-end sequencing (2x 100 bp). Sequences were aligned to the BN reference sequence Rno5 and compared to previous sequenced DA genomes15,16. Visual inspection of DAMut sequence alignments in Integrative Genomics Viewer (IGV), Broad Institute, revealed a region of partial or improperly paired alignments in intron 1 of the Sh3gl1 gene and indicated an insertion. The reads aligned in the region and their paired sequences were assembled using Trinity46. The inserted sequence was identified as an LTR/ERV1 element, using RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) prediction of the assembled sequences. The insertion was confirmed by Sanger sequencing.
Colony-forming units in kidneys of S. aureus injected mice
Kidneys were aseptically dissected, kept on ice, homogenized, serially diluted in PBS and spread on blood agar plates. After incubation for 24 h at 37 °C, the number of colony-forming units (CFU) per kidney pair was determined.
Tumor model
B16F10 melanoma cells were obtained from ATCC and cultured in 5% CO2 at 37 °C using 10% deactivated FBS in high glucose DMEM media supplemented with 4.5 grams/ml of glucose, 1 mM pyruvate, 1.5 g of NaHCO3/ 500 ml (Gibo GlutaMAX™, Thermofisher). Cells were washed two times with 37 °C PBS, trypsined with 1X EDTA trypsin, then neutralized with 1:1 ratio of 10% FBS DMEM growth medium to trypsin and 200 µl/mouse of serum free cell growth medium containing 1 × 106 B16F10 (passage 10) p10 melanoma cells were injected subcutaneously. Tumor volume was assessed using an electronic caliper and caliper formula (Length × Width2 × 0.52), where the width was the smallest dimension. Maximum allowed tumor size is 1.5cm2 as calculated by (Length × Width2 × 0.52).
Generation of Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 – SH3GL1 knock-out Jurkat cells
SH3GL1 CRISPR knock-out cells were generated and bought from GenScript Ltd. Two clones T3-5 and T3-21 were predicted to be SH3GL1 knock-outs and T3-23 to be wild type. The three clones were confirmed to be either SH3GL1 knock-out or wild-type Jurkat cells by and quantitative- polymerase chain reaction and western blotting (Supplementary Fig. 4a, b).
Proximity- ligase assay
Co-localization of EA2 and TCR was analysed by proximity ligation assay using the Duolink® PLA Starter Kit Mouse/Rabbit from Sigma (Cat. No DUO9210) according to the manufacturer’s description. EA2 was detected using primary rabbit anti-human EA2 (Sigma HPA021485) at 1ug/ml. TCR was detected using primary mouse anti-human CD3 (BD Biosciences Clone Hit3a) at 1ug/ml. Primary antibodies were detected using Duolink® In Situ PLA® Probe Anti-Rabbit PLUS, Affinity purified Donkey anti-Rabbit IgG (H + L) and Duolink® In Situ PLA® Probe Anti-Mouse MINUS, Affinity purified Donkey anti-Mouse IgG (H + L) provided by the kit according to manufacturer’s recommendations. Nuclei were stained using DAPI at 0.5ug/ml for 20 min in PBS. Cells were imaged on a Zeiss LSM 800 confocal laser scanning microscope (Zeiss). TexasRed was detected at λex594nm/λem624nm and DAPI at 358 nm/λem461nm. For image analysis, events were counted using the Analyze Particles function on ImageJ 1.52i. The number of TexasRed positive spots were related to the number of DAPI positive events to give a ratio of TexasRed positive events per cell.
Statistical analysis
Visualization and calculations of arthritis and EAE data and immunoassays was done using GraphPad Prism v5 software program. The non-parametrical Mann–Whitney U test was used for statistical evaluation of arthritis scoring data and immunoassay. P-values < 0.05 were considered significant.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data that support the findings of this study are available in the supplemented data source file and from the corresponding authors. Source data are provided with this paper.
References
Bos, W. H., van de Stadt, L. A., Sohrabian, A., Rönnelid, J. & van Schaardenburg, D. Development of anti-citrullinated protein antibody and rheumatoid factor isotypes prior to the onset of rheumatoid arthritis. Arthritis Res. Ther. 16, 405 (2014).
Siebert, S., Tsoukas, A., Robertson, J. & McInnes, I. Cytokines as therapeutic targets in rheumatoid arthritis and other inflammatory diseases. Pharmacol. Rev. 67, 280–309 (2015).
Okada, Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014).
Van Boxel, J. A. & Paget, S. A. Predominantly T-cell infiltrate in rheumatoid synovial membranes. N. Engl. J. Med. 293, 517–520 (1975).
Ranges, G. E., Sriram, S. & Cooper, S. M. Prevention of type II collagen-induced arthritis by in vivo treatment with anti-L3T4. J. Exp. Med. 162, 1105–1110 (1985).
Corthay, A., Johansson, A., Vestberg, M. & Holmdahl, R. Collagen-induced arthritis development requires alpha beta T cells but not gamma delta T cells: studies with T cell-deficient (TCR mutant) mice. Int. Immunol. 11, 1065–1073 (1999).
Sakaguchi, N. et al. Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nature 426, 454–460 (2003).
Milosevic, I. et al. Recruitment of endophilin to clathrin-coated pit necks is required for efficient vesicle uncoating after fission. Neuron 72, 587–601 (2011).
Baldassarre, T. et al. Endophilin A2 promotes TNBC cell invasion and tumor metastasis. Mol. Cancer Res. 13, 1044–1055 (2015).
So, C. W. et al. EEN encodes for a member of a new family of proteins containing an Src homology 3 domain and is the third gene located on chromosome 19p13 that fuses to MLL in human leukemia. Proc. Natl Acad. Sci. USA 94, 2563–2568 (1997).
Bäckdahl, L., Ekman, D., Jagodic, M., Olsson, T. & Holmdahl, R. Identification of candidate risk gene variations by whole-genome sequence analysis of four rat strains commonly used in inflammation research. BMC Genomics 15, 391 (2014).
Rintisch, C., Förster, M. & Holmdahl, R. Detection of arthritis‐susceptibility loci, including Ncf1, and variable effects of the major histocompatibility complex region depending on genetic background in rats. Arthritis Rheum. 60, 419–427 (2009).
Bergsteinsdottir, K., Yang, H. T., Pettersson, U. & Holmdahl, R. Evidence for common autoimmune disease genes controlling onset, severity, and chronicity based on experimental models for multiple sclerosis and rheumatoid arthritis. J. Immunol. 164, 1564–1568 (2000).
Vingsbo-Lundberg, C. et al. Genetic control of arthritis onset, severity and chronicity in a model for rheumatoid arthritis in rats. Nat. Genet. 20, 401–404 (1998).
Bäckdahl, L., Ekman, D., Jagodic, M., Olsson, T. & Holmdahl, R. Identification of candidate risk gene variations by whole-genome sequence analysis of four rat strains commonly used in inflammation research. BMC Genomics 15, 391 (2014).
Yau, A. C. Y. et al. Conserved 33-kb haplotype in the MHC class III region regulates chronic arthritis. Proc. Natl Acad. Sci. USA 113, E3716–E3724 (2016).
Maksakova, I. A. et al. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2, e2 (2006).
Santos-Rosa, H. et al. Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411 (2002).
Chahal, S. S., Matthews, H. R. & Bradbury, E. M. Acetylation of histone H4 and its role in chromatin structure and function. Nature 287, 76–79 (1980).
Nandakumar, K. S. & Holmdahl, R. Collagen antibody induced arthritis. Methods Mol. Med. 136, 215–223 (2007).
Holmberg, J. et al. Pristane, a non-antigenic adjuvant, induces MHC class II-restricted, arthritogenic T cells in the rat. J. Immunol. 176, 1172–1179 (2006).
Ringstad, N., Nemoto, Y. & De Camilli, P. The SH3p4/Sh3p8/SH3p13 protein family: binding partners for synaptojanin and dynamin via a Grb2-like Src homology 3 domain. Proc. Natl Acad. Sci. USA 94, 8569–8574 (1997).
Pizzolla, A. et al. Reactive oxygen species produced by the NADPH oxidase 2 complex in monocytes protect mice from bacterial infections. J. Immunol. 188, 5003–5011 (2012).
Bremell, T., Lange, S., Yacoub, A., Rydén, C. & Tarkowski, A. Experimental Staphylococcus aureus arthritis in mice. Infect. Immun. 59, 2615–2623 (1991).
Abdelnour, A., Bremell, T., Holmdahl, R. & Tarkowski, A. Clonal expansion of T lymphocytes causes arthritis and mortality in mice infected with toxic shock syndrome toxin-1-producing staphylococci. Eur. J. Immunol. 24, 1161–1166 (1994).
Boucrot, E. et al. Endophilin marks and controls a clathrin-independent endocytic pathway. Nature 517, 460–465 (2014).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 41, D991–D995 (2013).
Huppa, J. B., Gleimer, M., Sumen, C. & Davis, M. M. Continuous T cell receptor signaling required for synapse maintenance and full effector potential. Nat. Immunol. 4, 749–755 (2003).
Piotrowski, J. T., Gomez, T. S., Schoon, R. A., Mangalam, A. K. & Billadeau, D. D. WASH knockout T cells demonstrate defective receptor trafficking, proliferation, and effector function. Mol. Cell. Biol. 33, 958–973 (2013).
Liu, C.-Z. et al. Endophilin A2 influences volume-regulated chloride current by mediating ClC-3 trafficking in vascular smooth muscle cells. Circ. J. 80, 2397–2406 (2016).
Huang, E.-W. et al. Endophilin-A2-mediated increase in scavenger receptor expression contributes to macrophage-derived foam cell formation. Atherosclerosis 254, 133–141 (2016).
Borroto, A. et al. First-in-class inhibitor of the T cell receptor for the treatment of autoimmune diseases. Sci. Transl. Med. 8, 370ra184–370ra184 (2016).
Weiner, H. L. et al. Double-blind pilot trial of oral tolerization with myelin antigens in multiple sclerosis. Science 259, 1321–1324 (1993).
Sieper, J. et al. Oral type II collagen treatment in early rheumatoid arthritis. A double-blind, placebo-controlled, randomized trial. Arthritis Rheum. 39, 41–51 (1996).
Elliott, M. J., Feldmann, M. & Maini, R. N. TNF alpha blockade in rheumatoid arthritis: rationale, clinical outcomes and mechanisms of action. Int. J. Immunopharmacol. 17, 141–145 (1995).
Cohen, M. D. & Keystone, E. Rituximab for rheumatoid arthritis. Rheumatol. Ther. 2, 99–111 (2015).
Smolen, J. S. & Aletaha, D. Rheumatoid arthritis therapy reappraisal: strategies, opportunities and challenges. Nat. Rev. Rheumatol. 11, 276–289 (2015).
Winthrop, K. L. Infections and biologic therapy in rheumatoid arthritis: our changing understanding of risk and prevention. Rheum. Dis. Clin. North Am. 38, 727–745 (2012).
Bäcklund, J. et al. C57BL/6 mice need MHC class II Aq to develop collagen-induced arthritis dependent on autoreactive T cells. Ann. Rheum. Dis. 72, 1225–1232 (2013).
Lu, S., Carlsen, S., Hansson, A. S. & Holmdahl, R. Immunization of rats with homologous type XI collagen leads to chronic and relapsing arthritis with different genetics and joint pathology than arthritis induced with homologous type II collagen. J. Autoimmun. 18, 199–211 (2002).
Pizzolla, A., Wing, K. & Holmdahl, R. A glucose-6-phosphate isomerase peptide induces T and B cell-dependent chronic arthritis in C57BL/10 mice: arthritis without reactive oxygen species and complement. Am. J. Pathol. 183, 1144–1155 (2013).
Tuncel, J. et al. Animal models of rheumatoid arthritis (I): pristane-induced arthritis in the rat. PLoS ONE 11, e0155936 (2016).
Holmdahl, R. et al. Genetic analysis of mouse models for rheumatoid arthritis. Human Genome Methods 215, 215–238 (1998).
Broman, K. W., Wu, H., Sen, S. & Churchill, G. A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890 (2003).
Rintisch, C. et al. Natural variation of histone modification and its impact on gene expression in the rat genome. Genome Res. 24, 942–953 (2014).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Acknowledgements
We are very grateful for the SH3gl1 deficient mice kindly provided by Professor Pietro De Camilli at Yale University. We also thank Leonid Padyukov for providing human samples, Robert A Harris for critical review of manuscript and Uppmax for providing assistance in sequencing and computational infrastructure. The Laboratories for Chemical Biology at Karolinska Institutet (LCBKI) Hanna Axelsson, Martin Haraldsson, and Thomas Lundbäck for SH3GL1 assay development. This work was supported by grants from the Knut and Alice Wallenberg Foundation, the Swedish Association against Rheumatism, the Swedish Medical Research Council, the Swedish Foundation for Strategic Research. The research leading to these results has further received funding from the European Community’s IMI project BTCURE.
Funding
Open Access funding provided by Karolinska Institute.
Author information
Authors and Affiliations
Contributions
U.N. together with R.H. designed the overall study, performed, planned, analysed the data, and wrote the manuscript. C.R. performed and analysed the linkage analysis and ChIP sequencing and reviewed the manuscript. F.F., L.M., J.T., K.K., M.B., J.B., M.Y., K.S., M.B., M.Y.B., G.F.L., J.J., and L.B. performed experiments and reviewed the manuscript. D.E. performed the bioinformatic analyses and reviewed the manuscript. I.G. and N.H. provided advice and reviewed the manuscript. R.H. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks Michael Dustin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Norin, U., Rintisch, C., Meng, L. et al. Endophilin A2 deficiency protects rodents from autoimmune arthritis by modulating T cell activation. Nat Commun 12, 610 (2021). https://doi.org/10.1038/s41467-020-20586-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-020-20586-2
This article is cited by
-
The interplay between membrane topology and mechanical forces in regulating T cell receptor activity
Communications Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.