How nitrate regulates gene expression in legumes
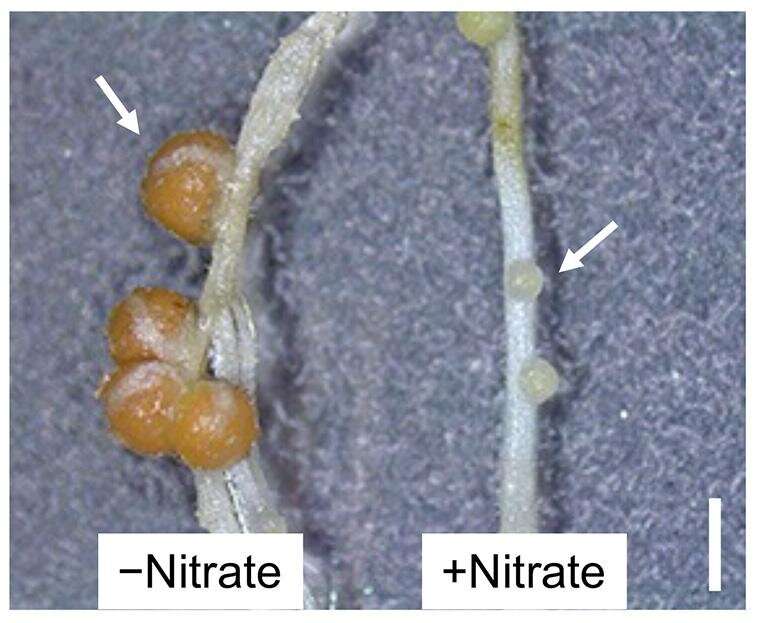
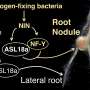
Plants in the bean family (legumes) form nodules on their roots to take up nitrogen. Legumes will stop nodule production when nitrogen is plentiful (Figure 1), but precisely how nitrate presence controls nodule formation in these plants has been a mystery. Now, researchers from Japan have found that interactions between proteins and nitrate can induce and repress genes, controlling nodulation with potential applications in sustainable agriculture.
In a study published in April in The Plant Cell, a research team from the University of Tsukuba has shown that the different DNA-binding properties between proteins that establish nodule development determine if genes involved in symbiosis that govern nodulation turn on or off and that this gene expression is nitrate-induced.
Until now, there was an incomplete understanding of the molecular activity determining how legumes stop nodulation in the presence of excess nitrate. Previous research identified transcription factors (proteins that help turn specific genes "on" or "off") involved with nodule formation, but that's just part of the story.
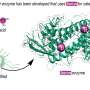
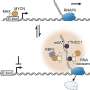
"Building on the previous identification of transcription factors for proteins (known as NLPs) involved in nodule inception, we sought to answer the question of how symbiotic gene expression facilitating nodulation is controlled by nitrate," says senior author of the study Professor Takuya Suzaki. "We tested specific NLPs and found that they have overlapping functions, causing nitrate-induced control of nodulation."
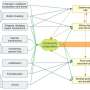
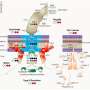
To examine these molecular interactions, the researchers analyzed RNA molecules and plant traits using proteins from Lotus japonicus. They found that some proteins have dual functions, acting as master regulators for nitrate-dependent gene expression. They also identified new protein binding sites and compared them to previously known ones. Their findings reveal basic principles relating to NLP-regulated transcription of symbiotic genes inhibiting nitrate nodulation.
The research team emphasized additional questions. Some NLPs are found in cell nuclei in response to nitrate and stop nodule production, while others constantly aggregate in nuclei irrespective of nitrate levels. For the latter, it is unclear how they function exclusively in the presence of nitrate. The location of the NLPs in the cell matters because translation (when RNA is coded into proteins) happens in the cell's cytoplasm. If changes to proteins occur after the genetic code has been read (post-translational modifications), it could explain how these NLPs access protein-protein interactions and regulate genes.
"Uncovering how transcription factors influence gene expression has been a missing piece to the puzzle of understanding plant transcription regulation," Professor Suzaki explains. "Our discoveries bring us closer to knowing what is possible within these complex molecular relationships, but there is plenty left to untangle. Future research should aim to answer the question of how nodulation is regulated by other NLPs and in other plant species of interest."
More information: Hanna Nishida et al. Different DNA-binding specificities of NLP and NIN transcription factors underlie nitrate-induced control of root nodulation, The Plant Cell (2021). DOI: 10.1093/plcell/koab103
Journal information: Plant Cell
Provided by University of Tsukuba