Abstract
Osteosarcoma (OS) is a type of bone tumor conferred with high metastatic potential. Attainable growth of tumors necessitates functional vasculature mediated by sprouting angiogenesis (SA) and intussusceptive angiogenesis (IA). However, the regulation of IA and SA is still unclear in OS. To understand the mechanisms adopted by OS to induce angiogenesis, initially, we assessed the expression profile of a set of miRNAs’ in both OS cells (SaOS2 and MG63) and normal bone cells. Amongst them, miR-432-5p was found to be highly downregulated in OS. The functional role of miR-432-5p in OS was further analyzed using miR-432-5p mimic/inhibitor. Platelet-derived growth factor-B (PDGFB) was found to be a putative target of miR-432-5p and it was further confirmed that the PDGFB 3′UTR is directly targeted by miR-432-5p using the luciferase reporter gene system. PDGFB was found to be secreted by OS to regulate angiogenesis by targeting the cells in its microenvironment. The conditioned medium obtained from miR-432-5p mimic transfected MG63 and SaOS2 cells decreased cell viability, proliferation, migration, and aorta ring formation in endothelial cells. The miRNA mimic/inhibitor transfected MG63 and SaOS2 cells were placed on SA (day 6) and IA (day 9) phase of CAM development to analyze SA and IA mechanisms. It was found that miR-432-5p mimic transfection in OS promotes the transition of SA to IA which was documented by the angiogenic parameters and SA and IA-associated gene expression. Interestingly, this outcome was also supported by the zebrafish tumor xenograft model. Corroborating these results, it is clear that miR-432-5p expression in OS cells regulates SA and IA by targeting PDGFB genes. We conclude that targeting miR-432-5p/PDGFB signaling can be a potential therapeutic strategy to treat OS along with other existing strategies.
Similar content being viewed by others
Introduction
Osteosarcoma (OS) is a high-grade malignant bone tumor that mainly affects children and adolescents with poor prognosis. Surgery and adjuvant chemotherapy are standard treatment modalities. However, the available chemotherapeutic drugs are known to cause fatal toxicity and deleterious side effects owing to off targeting problems [1]. No significant advancements in the therapeutic regimes for treating OS have been yet achieved. Exploring the molecular mechanisms associated with the pathophysiology of OS metastasis could offer new therapeutic targets for its treatment. Angiogenesis is the formation of new blood vessels that occurs by two distinct processes namely, sprouting angiogenesis (SA) and intussusceptive angiogenesis (IA). Deregulation of angiogenesis is one of the chief reasons for tumor growth and progression. Inhibition of tumor angiogenesis has become a popular anticancer strategy trailing behind chemotherapy. Unfortunately, the clinically approved antiangiogenic drugs are ineffective due to the activation of various compensatory mechanisms involving cascade of macromolecules which aids tumor angiogenesis. In-depth investigations into these compensatory pathways are essential for developing broad antiangiogenic therapies along with chemotherapy for attaining maximum benefits. Until now antiangiogenic therapies are focused on SA and often IA is overlooked due to the scarcity of experimental models.
OS growth and progression are the result of interactions between tumor cells with endothelial cells, tumor-associated macrophages, and smooth muscle cells [2]. Moreover, these interactions facilitate the progression of tumor growth by supplying numerous growth factors and cytokines such as platelet-derived growth factor (PDGF), transforming growth factor beta, endothelin-1, and angiotensin II [3]. Amongst them, PDGFB is a known regulator of tumor angiogenesis by activating downstream transcriptional factors and molecular signaling pathways [4, 5]. PDGFB potentially regulates vascular remodeling, maturation, and stability by recruiting pericytes and vascular smooth muscle cells (VSMC) to form nascent vascular networks. Depletion of PDGFB gene leads to early embryonic defect due to the reduction of pericytes and vascular leakiness [6]. Simultaneous overexpression of PDGFB in murine fibrosarcoma leads to the formation of high-density primitive vascular plexuses, neovascularization, growth, and metastasis of tumors [4]. The level of PDGFB produced by tumor cells governs the therapeutic effects of anti-PDGF drugs on vascular remodeling and metastasis [5].
There is plentiful evidence in the literature which signifies the involvement of microRNAs (miRNAs), a class of small, noncoding endogenous RNA molecules, in regulating tumor growth and progression. miRNAs are known to down regulate gene expression by targeting 3′UTR of specific mRNAs [7]. They regulate the status of various vascular remodeling diseases and targeting these miRNAs would lessen the severity of diseases. In PDGF signaling pathway, miR-4632 was found to target c-JUN and regulate the proliferation and apoptosis of PDGF-BB stimulated human pulmonary artery smooth muscle cells [8]. miR-221 induced by PDGF in primary VSMC significantly down regulates the targets c-Kit and p27Kip1 [9]. miR-26a contributes to the PDGFB-induced phenotypic switch of VSMC by targeting Smad1 [10]. With many evidences lined up emphasizing the involvement of miRNAs in regulating PDGF induced cellular changes, the discovery of new regulatory molecules involved in PGDF signaling holds great scientific and therapeutic interest. In the present study, we analyzed PGDF signaling and its regulatory molecules in the compensatory mechanisms of tumor angiogenesis and enlightened the role of SA and IA in OS microenvironment. This holds significant possibility for developing effective anticancer treatment regimens targeting angiogenesis.
Materials and methods
Cell culture and miRNAs transient transfection
The immortalized human OS cell lines (MG63, SaOS2, HOS) were purchased from NCCS, Pune, India, and human endothelial cell line (EA.hy926) was obtained as a kind gift from Dr. Suvro Chatterjee, Anna University, Chennai, India and Dr. C.J.S. Edgell, University of North Carolina, Chapel Hill, and were cultured in DMEM supplemented with 10% FBS (v/v) and 1% penicillin (w/v) and streptomycin (w/v) with standard condition, at 37 °C temperature and 5% CO2. MG63 is a human OS cell line derived from an OS of a 14-year-old male [11] and SaOS2 (Sarcoma osteogenic) is also an OS cell line derived from the primary OS of an 11-year-old Caucasian girl [12]. Another human OS cell line, HOS was derived from a 13-year-old Caucasian girl [13]. Initially, 50,000 of OS cells were seeded on 18 mm coverslips and incubated with the standard condition at 37 °C, 5% CO2 by adding 500 μl of media to reach 70% confluence. miRNAs transient transfection was carried out as described previously [14]. miR-432-5p mimic (50 nM) or miR-432-5p inhibitor (50 nM) or control miRNA (50 nM) (Applied Biosystems) was mixed with transfection reagent (X-treme Gene transfection reagent, Roche), and the transient transfection was performed as per the manufacturer’s instructions. miR-432-5p inhibitor is designated to bind with endogenous miR-432-5p, while miR-432-5p mimic is a similar sequence of endogenous miR-432-5p. miR-432-5p mimic/inhibitor was studied with a control miRNA. Upon transfection, the cells were incubated at standard conditions of 37 °C, 5% CO2 to reach 100% confluence. Similarly, silencer siRNA specific to PDGFB and non-silencer siRNA (scrambled siRNA) were purchased from Thermo Fisher Scientific. Transfection reagent (X-treme Gene transfection reagent, Roche) was mixed with siRNA-PDGFB (20 nM) or scrambled siRNAs (20 nM), and the transient transfection was performed as per the manufacturer’s instructions. Upon transfection, the cells were cultured for 24 h at standard conditions of 37 °C, 5% CO2.
Luciferase reporter assay
The wild 3′ UTR of PDGFB (sense: 5′ AAA CTA GCG GCC GCA TCT TCT TCC CTT AAC TCC AAG AT 3′, antisense: 3′ TTT GAT CGC CGG CGT AGA AGG GAA TTG AGG TTC TAG ATC 5′) and mutant 3′ UTR of PDGFB (sense: 5′ AAA CTA GCG GCC GCA TCT TCT TCC CTT AAC TCT TGA T 3′, antisense: 3′ TTT GAT CGC CGG CGT AGA AGG GAA TTG AGA ACT AGA TC 5′) were chemically synthesized. These sense and antisense primers of either wild or mutant PDGFB containing an internal NotI site were annealed and cloned between PmeI and XbaI restriction sites present in pmirGLO dual-luciferase miRNA target expression vector (Promega). The clones containing the PDGFB 3′UTR sequence were identified by NotI restriction enzyme. MG63 and SaOS2 cells were transiently transfected with miR-432-5p mimic/inhibitor or control miRNA (50 nM) along with wild/mutant PDGFB 3′UTR constructs (200 ng) using Lipofectamine 2000 transfection reagent (Invitrogen). After 24 h the luciferase activity was measured using dual-luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity [14,15,16].
Egg yolk angiogenesis assay
Preincubated white Leghorn chick (Gallus gallus) eggs were obtained from Government Poultry Station, Chennai. The standard conditions, 37 °C and 60% humidity were used to incubate the eggs. All assays were executed at Hamburger Hamilton (HH) stages 29 (also known as 6-day-old embryo) and at HH 35 (also known as 9-day-old embryo) embryos. The eggs were opened and released into prepared foam cups where the embryos were assured to face upwards and covered with a breathable polyethylene sheet. The shell-less embryos were incubated at 35.5 °C and 68% humidity [17]. In total, 18 mm coverslips seeded with OS cells, MG63 and SaOS2 with and without miRNA transfection were placed on the vascular bed so as the cells can directly be in contact with the embryos. Upon treatment for up to 6 h, the images were obtained using stereomicroscope equipped with Magnus digital camera and analyzed using AngioQuant software [18].
Real-time RT-PCR analysis
Total RNA was isolated with Trizol reagent (Invitrogen) as per the manufacturer’s protocol. cDNA was synthesized using Reverse Transcriptase kit as per the manufacturer’s instructions (Invitrogen). Real-time PCR analysis was executed using a SYBR premix Ex Taq II (TIi RNase H plus) (Takara). U6 and GAPDH primers were used to normalize miRNA and mRNA expression, respectively, for cell line study and β-actin primer was used for CAM assay-based mRNA expression. The primer sequences used for this study are tabulated in Supplementary Table 1. For mature miRNA detection, a micro Script II RT kit (Qiagen) was used for reverse transcription. Briefly, 1 µg template RNA, 1X miScript HiSpec buffer and 1X miScript Nucleics Mix were added and the volume was made-up to 20 µl of reverse transcription reaction mix using RNase free water. For expression, 1X QuantiTect SYBR Green PCR Master Mix, 1X miScript primer assay, 1X miScript Universal Primer, RNase free water, and template cDNA were added for the 50 µl reaction volume. The PCR thermal cycling profile was used as follows: 95 °C for 5 min as an initial denaturation step, followed by 40 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s, with a final extension of 72 °C for 5 min. The relative miRNA/mRNA expression level was calculated by ΔΔCt method of relative quantification [14].
Western blot analysis
Whole cell lysates were prepared using RIPA buffer containing protease inhibitor cocktail (Sigma-Aldrich) and 20 μg of total isolated protein was used for western blot analysis using the antibodies for PDGFB (1:1000) and CDK-2 (1:1000) (Santa Cruz Biotechnology). For Zebrafish, similarly, whole lysate was prepared from the embryos and western blot analysis was performed using the antibodies Ephrin-B2 (1:1000), Tie-2 (1:1000) (R&D Systems), Cald1 (1:1000) (Millipore), CXCR4 (1:1000), and α-tubulin (1:1000) (Genetex). Supersignal West Dura extended duration substrate (Thermo Scientific) was used to develop the signals of protein band. Western blot analysis was executed by following the protocol described elsewhere [14].
Chick aortic ring sprouting analysis
Chick aortic ring assay was executed as described elsewhere [19]. Initially, 50 μl of Matrigel was coated on coverslips and solidified for 30 min at 37 °C in a CO2 incubator. Chick aortic arches were removed from the heart of 12-day-old chick embryos and the aortic arches were then cut into small rings and placed on Matrigel-coated coverslips, and further 20 μl of Matrigel was again loaded on top of the aortic rings. The conditioned media was prepared from MG63, SaOS2 cells with and without miRNA transfection and incubated with aortic ring upto 36 h. After treatment the arches were washed using 1 × PBS, and imaged under an inverted microscope. The captured images were then processed for ring, junction, length, and size using AngioQuant software [18].
Cell proliferation and FDA
EA.hy926 cells were seeded at the concentration of 3 × 104/cm2 in 24-well plates and performed MTT (3,4,5-dimethylthiazol-2yl-2,5-diphenyl-2H-tetrazoliumbromide) assay as described previously [17]. Briefly, EA.hy926 cells were cultured in conditioned media prepared from MG63 and SaOS2 with and without miRNAs transfection. Cells were washed with PBS after 48 h of treatment and added with appropriate volume of MTT (5 mg/ml) solution followed by incubation at 37 °C for 4 h. After incubation, DMSO was used to replace MTT solution to dissolve the crystals formed. In the end, optical density was measured at 570 nm using a spectrophotometer. Similarly, after treatment, the cells were added with fluorescein diacetate (FDA) solution (30 μg/ml) and images were obtained under a fluorescent microscope with a ×20 objective [17].
Wound scratch assay
EA.hy926 cells (1 × 106 cells/ml) were seeded on 24-well plate and cultured upto 24 h to reach confluence. The monolayer cells were scratched with a 20 µl enzyme tip to make a linear wound and the cells were washed with 1x PBS and incubated with conditioned medium prepared from MG63 and SaOS2 cells with and without miRNA transfection. After 24 h, bright field images were taken with ×4 magnification under an inverted microscope. The rate of wound closure was calculated from the images using ImageJ software.
Boyden’s chamber-based migration assay
Boyden’s chamber assay was used to asses EA.hy926 cell’s migration potential under MG63 and SaOS2 cells conditioned media treatment. Boyden’s chamber contains two-chamber separated by collagen-coated polycarbonate membranes (8 μm pore size). EA.hy926 cells were loaded in the upper well and the lower well was filled with conditioned media prepared from MG63 and SaOS2 cells with and without miRNA transfection. The chambers were then allowed to incubate at 37 °C, 5% CO2 for 3 h and the cells were migrated across the membrane, and trapped to the lower part of the polycarbonate membrane. After the experiment, the membrane was fixed in 4% paraformaldehyde and then stained with 4′,6-diamidino-2-phenylindole and the fusion products were identified by fluorescence microscopy. EA.hy926 cell migration was calculated based on the number of migrated cells on the lower surface of the polycarbonate membrane. EA.hy926 cell number was counted before and after the experiment to calculate the proliferation rate of loaded cells in the Boyden’s chamber [20].
Human PDGFB quantification by ELISA kit
OS cell lines, MG63, SaOS2 cells were transfected with miR-432-5p mimic/inhibitor or control miRNA and cultured upto 24 h. The conditioned medium was prepared and subjected to PDGFB quantification by using human PDGFB ELISA Kit (Elabscience) as per the manufacturer’s instructions.
Animal model and tumor cell transplantation
Breeding and maintenance of zebrafish were carried out in house facility in accordance with established protocols [21]. The fishes were maintained at 28 °C with 10-h dark and 14-h light cycle. All protocols were approved by the institutional animal ethical committee of Anna University, Chennai, India. Initially, SaOS2 cells were transfected with control miRNA/miR-432-5p inhibitor/miR-432-5p mimic as described in the “Cell culture and miRNAs transient transfection” section. After 24 h of transfection, the cells were harvested and labeled with CellTracker™ Green CMFDA (5-chloromethylfluorescein diacetate) (Thermo Fisher) as per the manufactures’ protocol and implanted in embryos as described [22, 23]. Briefly, the prepared SaOS2 cells (~1000 cells) were implanted into 2 days postfertilization embryos. A glass needle loaded with ~10 nl of cell suspension solution and was injected into the yolk sac near to the duct of cuvier by Femtojet injector (Eppendorf). The grafted animals were allowed to recover at 28 °C for 1 h and maintained at 35 °C. After 24 h postimplantation the tumor cells were imaged using fluorescence microscopy.
Statistical analysis
All data were represented as mean ± standard deviation with a minimum of n = 3. The significant difference (p < 0.05) between groups was calculated by the Student’s t test. The statistical significance of the intergroup differences was analyzed using one-way analysis of variance followed by the followed by Tukey’s multiple comparison (multiple groups) and p < 0.05 was considered as statistically significant.
Results
Identification and expression of miRNAs targeting PDGFB in osteosarcoma
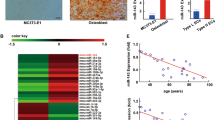
Understanding the signaling mechanisms prevailing in OS microenvironment will help us to identify potential markers involved in the development and progression of OS. Hence, we were interested to investigate the role of signaling molecules that are involved in regulation of tumor angiogenesis by OS. Initially, we analyzed the expression profile of PDGFB across all tumor samples and paired with normal tissues using a bioinformatics tool (http://gepia.cancer-pku.cn/detail.php) (Fig. S3). The results indicate the distributional expression of PDGFB in normal and tumor cells. There are literatures that showed the increased expression of PDGF in human OS clinical samples [24,25,26].
There is a differential expression of miRNAs between normal osteoblast and OS cells [27]. It has been found that 26 miRNAs were downregulated in OS cells compared to normal osteoblasts. Among them nine miRNAs (miR-29b-1-5p, miR-431-5p, miR-432-5p, miR-410-3p, miR-335-5p, miR-382-5p, miR-154-5p, miR-34a-3p, miR-136-5p) putatively target PDGFB 3′UTR (Fig. S1). Additionally, based on bioinformatics analysis, the thermodynamic stability interaction between miRNAs and PDGFB was tabulated and three out of nine miRNAs (miR-432-5p, miR-34a-3p, miR-410-3p) indicated higher miRmap score, which shows the higher possibility for direct targeting (Fig. S2). Amongst these three miRNAs, miR-432-5p possess higher negative miRmap score and thermodynamic stability of miR-432-5p interaction with PDGFB and is shown in the right side of Fig. S1. Based on miRmap target score from Fig. S2, miR-432, miR-410 and miR-34a were selected for further expression analysis. Total RNA was isolated from OS cells (MG63, SaOS2), 14 days differentiated human osteoblast cells and rat calvarial cells and was subjected to real-time RT-PCR analysis. The results indicated that these miRNAs are downregulated in OS cells compared to normal osteoblasts (Fig. 1). The results are similar and align with a previously published data [27].
Total RNA was isolated from OS cell line, MG63, SaOS2, differentiated human osteoblast and rat calvarial cells and subjected to pre-miRNAs expression using specific primers. The relative expression of these pre-miRNAs was calculated after normalization with U6 expression. (*) indicates significant decrease compared to normal osteoblast. Results are expressed as the mean ± SD. *Indicates significantly decreased compare to differentiated human osteoblast and rat calvarial cells and analyzed by one-way ANOVA followed by Tukey’s multiple comparison.
miR-432-5p targeted and suppressed the expression of PDGFB
We speculated that miR-432-5p could be involved in the angiogenesis regulation of OS cells as observed by its expression in Fig. 1 and the bioinformatics prediction also revealed that the PDGFB 3′UTR sequence has putative miR-432-5p binding sites as shown in Fig. S1. Hence, to validate the putative target genes of miR-432-5p, MG63 cells were transiently transfected with miR-432-5p mimic/inhibitor or control miRNA, followed by total RNA isolation and real-time RT-PCR analysis. The results demonstrated that after miR-432-5p mimic transfection the mRNA expression level of PDGFB was significantly decreased in MG63 and SaOS2 cells (Fig. 2A). As anticipated, the protein expression of PDGFB was consistent with that of mRNA expression levels (Fig. 2B, C).
miR-432-5p was selected for further functional analysis. miR-432-5p mimic, inhibitor, and control miRNAs were transfected to MG63 and SaOS2 cells to study its target gene, PDGFB expression. A Total RNA was isolated and real-time RT-PCR was carried out using the primers for PDGFB genes. Fold change of mRNAs was calculated after normalization with GAPDH. B After transfection, whole cell lysates were prepared and subjected to Western blot analysis using the antibodies as shown. CDK-2 was used as a loading control. C Densitometry scanning of these proteins expression by Western blot after normalization with CDK-2. D ELISA-based quantification of PDGFB secreted in the medium by MG63 and SaOS2 cells represented as relative fold induction. Results are expressed as the mean ± SD. *Indicates significantly decreased compared to control miRNA or/and inhibitor. It was analyzed by one-way ANOVA followed by Tukey’s multiple comparison.
Additionally, the conditioned medium from MG63 and SaOS2 cells transfected with miR-432-5p mimic/inhibitor or control miRNA was quantified for PDGFB using ELISA kit (Fig. 2D). A similar trend in PDGFB levels was observed and consistent with its mRNA and protein levels. Next, the intracellular level of miR-432-5p was analyzed in SaOS2 cells transfected with miR-432-5p mimic/inhibitor or control miRNA. The expression level of miR-432-5p was significantly increased in mimic transfection, while it was decreased in inhibitor transfection compared to control miRNA (Fig. S4).
Further, to confirm experimentally that PDGFB is a direct target of miR-432-5p, a dual-luciferase reporter system was employed. The pmirGLO construct (Promega), having the Renilla and firefly luciferase genes was used for cloning of PDGFB 3′ UTR target region. As shown in Fig. 3A, wild or mutant PDGFB 3′ UTR sequences were chemically synthesized, annealed, and cloned into pmirGLO vector. The Renilla luciferase was used for normalization of the firefly luciferase activity in pmirGLO vector as it is constitutively active. The pmirGLO vector containing the wild/mutant PDGFB 3′ UTR downstream to the firefly luciferase reporter gene was co-transfected with miR-432-5p mimic/inhibitor or control miRNA in MG63 (Fig. 3B) and SaOS2 (Fig. 3C) cells. After 24 h of transfection, the luciferase activity was analyzed. The results depicted that luciferase activity was significantly decreased in the cells co-transfected with the pmirGLO construct containing PDGFB wild 3′ UTR along with the miR-432-5p mimic, whereas control miRNA or miR-432-5p inhibitor showed no significant alteration in the luciferase activity (Fig. 3B, C). The mutant 3′ UTR of PDGFB pmirGLO construct was not altered after transfections with control miRNA, miR-432-5p mimic or inhibitor. The results demonstrate the specificity of the 3′ UTR of PDGFB to miR-432-5p in OS cells. Thus, this experiment suggested that miR-432-5p directly targets the 3′ UTR of the PDGFB gene in OS, MG63, and SaOS2 cells. However, the other putative targets of miR-432-5p predicted by TargetScan (http://www.targetscan.org/vert_72/) cannot be ruled out.
A A part of PDGFB 3′UTR region was putatively predicted as a target of miR-432-5p (https://mirmap.ezlab.org/). The putatively predicted target sequence of PDGFB and its mutated sequence were cloned/inserted into pmirGLO construct. MG63 (B) and SaOS2 (C) cells were transiently co-transfected pmirGLO construct (containing either wild/mutant sequence of PDGFB 3′ UTR) with miR-432-5p mimic/inhibitor or control miRNA. After 24 h, relative luciferase activity was measured by normalization of firefly luciferase activity with Renilla luciferase activity. Results are expressed as the mean ± SD. *Indicates significantly decreased luciferase activity and it was analyzed by one-way ANOVA followed by Tukey’s multiple comparison.
miR-432-5p/PDGFB signaling regulation in osteosarcoma regulates endothelial cells proliferation, migration, and sprouting
We found that miR-432-5p suppresses PDGFB expression in OS cells (Figs. 1–3). Hence, to determine whether miR-432-5p/PDGFB signaling in OS has an influence on tumor angiogenesis, the endothelial cells proliferation, migration, and sprouting were analyzed. Initially, MG63 and SaOS2 cells were transiently transfected with control miRNA or miR-432-5p mimic/inhibitor and after 24 h, the conditioned medium was prepared and incubated with endothelial cells (EA.hy926 cells) to check its proliferation, migration, and sprouting. EA.hy926 cells were cultured with conditioned medium up to 48 h and cell proliferation was assessed by MTT assay and viability was measured by FDA staining (Fig. 4A, B). Also, EA.hy926 cells proliferation was assessed by MTT assay after 24 h of treatment with conditioned medium prepared from HOS cells transfected with mimic/inhibitor or control miRNA (Fig. S5A). The results indicate that the conditioned medium from miR-432-5p mimic transfected OS cells significantly reduced endothelial cell proliferation without altering viability compared to the conditioned medium from control miRNA or miR-432-5p inhibitor transfection. Similarly, the other phenotype of angiogenesis, endothelial cell migration was assessed by wound scratch assay (Figs. S6 and 4C) and Boyden’s chamber migration assay (Figs. 4B and S5B). The results indicated a significant reduction in endothelial cell migration, wound closure, and cell migration index upon treatment with the conditioned medium from miR-432-5p mimic compared to conditioned medium from control miRNA or miR-432-5p inhibitor transfection. In addition to this, chick aortic ring sprouting analysis was performed to understand the endothelial sprouting. The aortic ring was prepared and treated with conditioned medium of miR-432-5p mimic/inhibitor or control miRNA control transfected MG63 or SaOS2 cells (Fig. 5A–C). Angiogenic parameters including number of rings, length, and tubule size, and number of junctions were analyzed after treatment wherein significant reduction was observed upon miRNA mimic transfection compared to inhibitor or control miRNA transfection. Expression of PDGFB in OS cells may regulate tumor angiogenesis, which is documented by analyzing these parameters. Furthermore, to check the possibility of any direct effect of miR-432-5p on endothelial cells’ phenotypes, the EA.hy926 cells were treated with the conditioned medium of OS cell lines, SaOS2 and HOS cells transfected with miR-432-5p mimic/inhibitor or control miRNA. After 24 h, total RNA was isolated from EA.hy926 cells and the expression of miR-432-5p was analyzed. The result showed that there was no significant change in miR-432-5p expression in mimic/inhibitor or control miRNA transfection compared to naive endothelial cells (without any treatment) (Fig. S7). The results suggest that miR-432-5p has no direct effect on endothelial cells.
Conditioned medium was prepared from miR-432-5p mimic/inhibitor transfected MG63 and SaOS2 cells and treated with EA.hy926 cells and cell proliferation (A) and viability (B) was evaluated. FDA staining was carried out for analyze cell morphology. C miR-432-5p/PDGFB signaling regulation in OS regulates endothelial cells migration. Conditioned medium was prepared from miR-432-5p mimic transfected MG63 and SaOS2 cells and treated with EA.hy926 cells and cell migration was evaluated. C EA.hy926 cell migration was calculated from scratch wound assay, Fig. S6. D shows cell migration evaluated by Boyden chamber. Results are expressed as the mean ± SD. *Indicates significantly decreased compared to control and/or inhibitor and (#) indicates significant increase in area of wound closure compared to control miRNA after 24 h. It was analyzed by one-way ANOVA followed by Tukey’s multiple comparison.
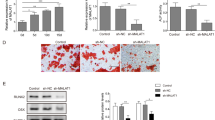
A 12-day-old chick embryo aortas were cut into small pieces. The aortas were placed between the Matrigel, and provided conditioned media prepared from MG63 and SaOS2 cells transfected with miR-432-5p mimic/inhibitor or control miRNA. Angiogenic parameters such as number of rings, length, size of the tubule, and number of junctions were quantified and plotted. B The ring and junction number was quantified under treatment compared with control miRNA transfection. C Similarly, the length and size were measured and plotted the graph. Results are expressed as the mean ± SD. *Indicates significantly decreased compared to control and/or inhibitor. It was analyzed by one-way ANOVA followed by Tukey’s multiple comparison.
miR-432-5p/PDGFB signaling regulation in osteosarcoma promotes intussusceptive angiogenesis
Recently, we developed a unique model to screen the angiogenic induction of tumor cells using chick embryo [17]. To perform egg yolk angiogenesis assay using OS cell lines, MG63 and SaOS2 cells were cultured on coverslips. Different numbers (500, 5000, and 50,000 cells) of MG63 and SaOS2 cells were cultured on coverslips and used for the initial screening (Fig. S8A–D). After 6 h of treatment with coverslip containing MG63 and SaOS2 on the CAM vascular bed, the images were captured and subjected to AngioQuant software to document the changes in angiogenic parameters. The software is used to detect the rate of angiogenesis in a specified field of the image by measuring the size, length, and number of junctions present in the blood vessels. Although all these three parameters are important to symbolize the rate of angiogenesis, the fold change in number of junctions of the blood vessels is a key indicator as it denotes the formation of new blood vessels. The results showed that a similar trend of angiogenic effect was observed between the OS cells in different numbers. However, for further confirmation, MG63 and SaOS2 cells were compared with primary rat calvarial cells (osteoprogenitor cells) in egg yolk angiogenesis assay (Fig. S9). The blood vessel size, length, and junction were significantly increased in OS cell lines, MG63 and SaOS2 cells treatment compared to primary osteoblasts.
To understand miR-432-5p/PDGFB signaling regulation in OS regulated tumor angiogenesis, the CAM model was used. During the development of chick embryo, CAM vascularization undergoes sprouting and intussusceptive phases for angiogenesis. From days 5 to 7, the CAM development is considered as the sprouting phase of angiogenesis. In this phase, the system prefers for capillary network growth. From days 8 to 12 is considered as an intussusceptive phase, where SA is switched into IA [17]. In our previous study, the angiogenesis pattern during CAM development from days 5 to 13 was analyzed. The CAM vasculature was imaged and the angiogenic parameters were analyzed using Angioquant [28]. Based on the angiogenic parameters, day 6 is ideal for SA analysis and day 9 is optimal for IA analysis. Hence, in the present study, we used days 6 and 9 CAM developmental stages to understand miR-432-5p/PDGFB signaling regulation of IA in OS.
MG63 and SaOS2 cells were cultured in coverslip and transiently transfected with miR-432-5p mimic/inhibitor or control miRNAs and after 24 h the coverslip containing OS cells were inversely placed on CAM vasculature at days 6 and 9 (Figs. 6 and 8). The angiogenic parameters were quantified using Angioquant software and compared between 2 and 6 h time interval. The result indicated that both at SA and IA phase of CAM vasculature miR-432-5p mimic transfected MG63 and SaOS2 cells treatment showed decreased blood vessel size, length, and junction (Figs. 6 and 8) compared with control miRNA or miR-432-5p transfected cells. Additionally, SA or IA-associated markers expression was analyzed by real-time RT-PCR analysis to understand miR-432-5p/PDGFB signaling in OS regulation of SA or IA.
MG63 and SaOS2 cells containing coverslip were transfected with miR-432-5p minic/inhibitor or control miRNA and inversely placed on sprouting phase (day 6) of CAM. After treatment, CAM images were obtained at 2 and 6 h using stereomicroscope and analyzed by Angiquant software. The formation of blood vessel was denoted by fold change of junction, length, and size. Results are expressed as the mean ± SD. *Indicates significantly decreased compared to control and/or inhibitor. It was analyzed by one-way ANOVA followed by Tukey’s multiple comparison.
MG63 and SaOS2 cells were cultured on coverslips and transfected with miR-432-5p mimic/inhibitor or control mRNAs and placed on CAM vasculature at days 6 and 9. After 6 h of treatment, total RNA was isolated from CAM bed and subjected to real-time RT-PCR analysis for Ephrin-B2, Tie-2 (SA-associated), TEM8, CALD1, CXCR4 (IA-associated) mRNAs (Figs. 7 and 9). SA and IA markers expression was analyzed during CAM development from days 5 to 13. The result confirms that Ephrin-B2 and Tie-2 are reported as SA-associated genes, while TEM8, CALD1, and CXCR4 are associated with IA [17]. The markers expression analysis indicated that the IA-associated genes, TEM8, CALD1, CXCR4 mRNAs expression were decreased in MG63 and SaOS2 cells transfected with miR-432-5p mimic compared to control miRNA or miR-432-5p inhibitor at both SA phase and IA phase of CAM. Moreover, the SA-associated markers, Ephrin-B2, Tie-2 mRNAs was not significantly altered by the treatment at both SA and IA phase of CAM. These results indicate that miR-432-5p/PDGFB signaling in OS may regulate IA during tumor development.
Six days old embryos’ CAM were exposed with MG63 or SaOS2 cells containing coverslip and after 6 h of treatment the bed was excised and subjected to real-time RT-PCR analysis. TEM8, CALD1, and CXCR4 are selected IA-associated genes, while Tie-2 and Ephrin-B2 are selected SA-associated genes. β-actin mRNA expression was used to normalize these mRNAs expression. Results are expressed as the mean ± SD. *Indicates significantly decreased compared to control and/or inhibitor. It was analyzed by one-way ANOVA followed by Tukey’s multiple comparison.
MG63 and SaOS2 cells containing coverslip were transfected with miR-432-5p minic/inhibitor or control miRNA and inversely placed on intussusceptive phase (day 9) of CAM. After treatment, CAM images were obtained at 2 and 6 h using stereomicroscope and analyzed by Angiquant software. The formation of blood vessel was denoted by fold change of junction (A), length (B), and size (C). Results are expressed as the mean ± SD. *Indicates significantly decreased compared to control and/or inhibitor. It was analyzed by one-way ANOVA followed by Tukey’s multiple comparison.
Nine days old embryos of CAM were treated with miR-432-5p mimic/inhibitor or control miRNA transfected MG63 or SaOS2 cells containing coverslip. After 6 h of treatment, the bed was excised and performed real-time RT-PCR analysis for selected IA-associated (TEM8, CALD1, CXCR4) and SA-associated (Tie-2, Ephrin-B2) markers expression. β-actin mRNA expression was used to normalize these mRNAs expression. (*) denotes significant decrease compared to control. Results are expressed as the mean ± SD. *Indicates significantly decreased compared to control and/or inhibitor. It was analyzed by one-way ANOVA followed by Tukey’s multiple comparison.
Furthermore, it is essential to understand whether the release of PDGFB by OS cells regulates angiogenesis. Various reports showed the involvement of PDGFB in OS and its regulation in tumor angiogenesis. For instance, down regulation of PDGFB by shRNA in breast cancer cells significantly reduced tumor angiogenesis [29]. PDGF-BB promotes tumor growth and metastasis through stimulation of angiogenesis and blocking the PDGFB signaling by imatinib inhibitor or blocking antibody inhibits vascular mimicry and tumor growth [30, 31]. Overexpression of PDGFB increases the growth, invasion, and angiogenesis of gastric carcinoma [32]. However, we inhibited PDGFB expression in OS cells by siRNA transfection and analyzed its effect on angiogenesis. Initially, MG63 and SaOS2 cells were transfected with scrambled siRNA and/or siRNA specific to PDGF and after 24 h the cells were subjected to total protein extraction and subjected to ELISA assay to check the level PDGFB silencing (Fig. S10A). The PDGFB expression in siRNA-PDGFB transfected cells was significantly decreased compared to scramble siRNA transfection. Similarly, MG63 and SaOS2 cells were cultured on coverslips and transfected with scrambled siRNA and/or siRNA-PDGFB and after 24 h, the coverslips containing OS cells were inversely placed on CAM vasculature at day 6 (Fig. S10B). The angiogenic parameters were quantified using Angioquant software and compared between 2 and 6 h time intervals. Reduction in blood vessel size, length, and junction was observed in siRNA-PDGFB transfected OS compared to scramble siRNA transfection. This result supports that PDGFB released from OS cells regulates tumor angiogenesis. Overall, the results highlight that OS cells enhance IA mechanism through PDGFB signaling and its inhibition by miR-432-5p controls OS angiogenesis. Therefore, screening and targeting miR-432-5p/PDGFB signaling along with other bioactive molecules could be an improved therapeutic strategy to inhibit IA in OS.
miR-432-5p/PDGFB signaling regulation in osteosarcoma promotes intussusceptive angiogenesis in vivo
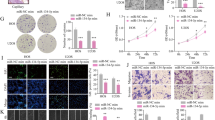
We were then interested to assess role of miR-432-5p/PDGFB signaling in OS regulation of IA using a zebrafish model. The zebrafish tumor xenograft model used in the study is based on the implantation of human tumor cells in the embryos [22, 23]. To assess the miR-432-5p/PDGFB signaling in IA, SaOS2 cells were initially transfected with miRNAs, labeled with CellTracker™ Green CMFDA, and injected into zebrafish embryo at 2 days postfertilization. The protocol used for tumor cell implantation in zebrafish is depicted in Fig. 10A. The grafted tumor cells were easily tracked with the help of CellTracker™ Green CMFDA labeling. After 24 h of implantation, successful homing of tumor cells around the gastrointestinal area is seen which confirms tumor presence (Fig. 10B). The survival rate of zebrafish embryos after microinjection was recorded for upto 6 days and there was around 60% survival with tumor cell injection compared to control (1× PBS) injection. However, no difference in survival rate was observed between miRNAs transfection in tumor cells (Fig. 10C). To determine the miR-432-5p/PDGFB signaling in IA, after 24 h of OS cell implantation in zebrafish embryo, the SA and IA markers expression was analyzed using western blot analysis (Fig. 10D). IA-associated markers; CXCR4 and CALD1 protein expression was decreased in fishes implanted with miR-432-5p mimic transfected SaOS2 cells compared to control miRNA or miR-432-5p inhibitor. Surprisingly, the SA-associated markers; Ephrin-B2 and Tie-2 protein expression were not altered. Therefore, the in vivo assessment indicates that miR-432-5p/PDGFB signaling in OS may regulate IA during tumor development.
A A schematic illustration of osteosarcoma cell implantation in zebrafish embryo. CellTracker™ Green CMFDA labeled SaOS2 cells were implanted into 2 dpf zebrafish embryos, which were screened after 24 h. B Survival rate of zebrafish embryos was observed until 6 dpf. 1XPBS injected embryos were considered as control group (n = 30 animals per group). C miRNA transfected SaOS2 cells were labeled with CellTracker™ Green CMFDA and injected into 2 dpf zebrafish embryos and after 24 h, the injected cells in embryos were tracked using fluorescent microscope (×10 magnification). D After 24 h of tumor cell implantation, whole lysate was prepared from embryo and performed western blot analysis using the antibodies as shown. α-tubulin was used as an internal loading control.
Discussion
OS is reported to have high metastatic potential and associated with poor patient survival rate [2]. However, the complex relationship existing between OS and its microenvironment is yet to be unraveled. Autocrine and paracrine secretions play a vital role in establishing cell to cell communication in tumor microenvironment [33]. These factors facilitate in establishing angiogenesis to promote tumor growth and provide metastatic potential [34, 35].
Among several growth factors reported to be involved in angiogenesis, vascular endothelial growth factors (VEGF), fibroblast growth factor-2, and PDGF are considered to be vital. PDGF is produced from both tumor and stromal cells and the accumulating evidences portray the importance of PDGF signaling in angiogenesis progression under both normal and tumor conditions [34, 36]. In OS and fibrosarcoma, tumor cells co-express PDGF and its receptor which leads to an increased rate of tumor progression by autocrine regulation [37]. Secretion of PDGFB by melanoma increases the number of pericytes within the tumor microenvironment and shows an increased tumor progression rate [38]. PDGFR inhibitor inhibits tumor PDGFRβ phosphorylation and also inhibits PDGFB-induced angiogenesis which results in inhibition of tumor development [39]. Generally, these expressions are cytoplasmic, and other than the tumor, they are also found in small blood vessels and scattered cells within tumor and its surrounding microenvironment.
In our study, by using a bioinformatics analysis, we found the distributional expression of PDGFB in both normal and tumor cells. It requires a convincing correlation between angiogenesis mechanism and PDGF produced by OS cells. The reports showed that PDGFB regulates the induction of angiogenesis but the mechanism by which PDGFB induces angiogenesis is not fully understood. It is hypothesized that the effects could be accomplished directly by the activation of PDGF signaling and/or indirectly, thereby increasing the amount of stromal VEGF production.
It is essential to propose alternative strategies to predict the prognosis and treatment in OS. miRNAs are highly involved in cancer pathogenesis and could serve as potential biomarkers. The evidence indicates that several tumors including OS are tightly regulated by miRNAs [7, 14]. Based on the thermodynamic stability and miRmap target score we selected three miRNAs that putatively target PDGFB. Amongst them, we found that miR-432-5p directly targeted PDGFB.
There is a possibility that miR-432-5p may regulate the release of other angiogenesis factors by PDGFB independent mechanisms. Angiogenesis involves proliferation, migration, and tube formation of endothelial cells to form new blood vessels [40]. Tumor microenvironment prefers IA to compensate for the demand when there is no enough resource to establish SA in a short time. IA is critically involved in hyper-proliferative diseases like a tumor, hypoxic mouse retina, liver cirrhosis, nephritis, and inflammation [41], In this study we found that the conditioned medium from miR-432-5p mimic transfected OS cells decreased endothelial cell proliferation, migration, and endothelial sprouting compared to miR-432-5p inhibitor or control miRNA transfection. Hence, we can conclude that miR-432-5p suppresses angiogenesis through PDGFB-dependent expression in human OS cells. In this study, we also confirmed that miR-432-5p/PDGFB signaling possess influence on tumor angiogenesis by limiting endothelial cells proliferation and sprouting in an in house chick embryo model.
A reasonable size of development of tumor necessitates the functional vasculature by two distinguished processes (1) SA develops new vessels from preexisting ones and (2) IA extends the inner vessel wall into the lumen of the blood vessel to split a single vessel into two [42]. Hence, IA is different from SA because it has no necessary requirement for endothelial cell proliferation and vascular remodeling. In this study, supported by the concomitant increase in IA-associated gene expression over SA both in vitro and in vivo, we conclude that miR-432-5p/signaling regulated IA over SA during tumor development.
Overall, to conclude the present study, we found that pre-miR-432 expression was decreased in OS and functional analysis showed that miR-432-5p directly targets PDGFB. miR-432-5p regulation of PDGFB signaling in OS suppresses tumor angiogenesis which was documented by decreased endothelial cells proliferation, migration, aorta ring formation assay. Additionally, miR-432-5p/PDGFB signaling in OS regulates tumor angiogenesis by switching from SA to IA which was documented by chick embryo and zebrafish models. Overall, we speculate that targeting miR-432-5p/PDGFB signaling pathway along with other molecules could be a better therapeutic option in controlling OS microenvironment contributed tumor angiogenesis, by specifically targeting SA and IA mechanisms.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Zhang Y, Yang J, Zhao N, Wang C, Kamar S, Zhou Y, et al. Progress in the chemotherapeutic treatment of osteosarcoma. Oncol Lett. 2018;16:6228–37.
Xie L, Ji T, Guo W. Anti-angiogenesis target therapy for advanced osteosarcoma (review). Oncol Rep. 2017;38:625–36.
Ng CF, Frieboes HB. Model of vascular desmoplastic multispecies tumor growth. J Theor Biol. 2017;430:245–82.
Nissen LJ, Cao R, Hedlund E-M, Wang Z, Zhao X, Wetterskog D, et al. Angiogenic factors FGF2 and PDGF-BB synergistically promote murine tumor neovascularization and metastasis. J Clin Invest. 2007;117:2766–77.
Xue Y, Lim S, Yang Y, Wang Z, Jensen LDE, Hedlund E-M, et al. PDGF-BB modulates hematopoiesis and tumor angiogenesis by inducing erythropoietin production in stromal cells. Nat Med. 2011;18:100–10.
Lindahl P, Hellström M, Kalén M, Karlsson L, Pekny M, Pekna M, et al. Paracrine PDGF-B/PDGF-Rbeta signaling controls mesangial cell development in kidney glomeruli. Dev Camb Engl. 1998;125:3313–22.
Vimalraj S, Miranda PJ, Ramyakrishna B, Selvamurugan N. Regulation of breast cancer and bone metastasis by microRNAs. Dis Markers. 2013;35:369–87.
Qian Z, Li Y, Chen J, Li X, Gou D. miR-4632 mediates PDGF-BB-induced proliferation and antiapoptosis of human pulmonary artery smooth muscle cells via targeting cJUN. Am J Physiol Cell Physiol. 2017;313:C380–91.
Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J Biol Chem. 2009;284:3728–38.
Yang X, Dong M, Wen H, Liu X, Zhang M, Ma L, et al. MiR-26a contributes to the PDGF-BB-induced phenotypic switch of vascular smooth muscle cells by suppressing Smad1. Oncotarget. 2017;8:75844–53.
Billiau A, Edy VG, Heremans H, Van Damme J, Desmyter J, Georgiades JA, et al. Human interferon: mass production in a newly established cell line, MG-63. Antimicrob Agents Chemother. 1977;12:11–15.
Fogh J, Fogh JM, Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J Natl Cancer Inst. 1977;59:221–6.
McAllister RM, Nelson-Rees WA, Peer M, Laug WE, Isaacs H, Gilden RV, et al. Childhood sarcomas and lymphomas. Characterization of new cell lines and search for type-C virus. Cancer. 1975;36:1804–14.
Vimalraj S, Partridge NC, Selvamurugan N. A positive role of microRNA-15b on regulation of osteoblast differentiation. J Cell Physiol. 2014;229:1236–44.
Vimalraj S, Selvamurugan N. Regulation of proliferation and apoptosis in human osteoblastic cells by microRNA-15b. Int J Biol Macromol. 2015;79:490–497.
Vishal M, Vimalraj S, Ajeetha R, Gokulnath M, Keerthana R, He Z, et al. MicroRNA-590-5p stabilizes Runx2 by targeting Smad7 during osteoblast differentiation. J Cell Physiol. 2017;232:371–80.
Vimalraj S, Bhuvaneswari S, Lakshmikirupa S, Jyothsna G, Chatterjee S. Nitric oxide signaling regulates tumor-induced intussusceptive-like angiogenesis. Microvasc Res. 2018;119:47–59.
Niemistö A, Dunmire V, Yli-Harja O, Zhang W, Shmulevich I. Robust quantification of in vitro angiogenesis through image analysis. IEEE Trans Med Imaging. 2005;24:549–53.
Siamwala JH, Veeriah V, Priya MK, Rajendran S, Saran U, Sinha S. Nitric oxide rescues thalidomide mediated teratogenicity. Sci Rep. 2012;2:679.
Tamilarasan KP, Kolluru GK, Rajaram M, Indhumathy M, Saranya R, Chatterjee S. Thalidomide attenuates nitric oxide mediated angiogenesis by blocking migration of endothelial cells. BMC Cell Biol. 2006;7:17.
Avdesh A, Chen M, Martin-Iverson MT, Mondal A, Ong D, Rainey-Smith S, et al. Regular care and maintenance of a zebrafish (Danio rerio) laboratory: an introduction. J Vis Exp. 2012:69;e4196.
Nicoli S, Presta M. The zebrafish/tumor xenograft angiogenesis assay. Nat Protoc. 2007;2:2918–23.
Chiavacci E, Rizzo M, Pitto L, Patella F, Evangelista M, Mariani L, et al. The zebrafish/tumor xenograft angiogenesis assay as a tool for screening anti-angiogenic miRNAs. Cytotechnology. 2015;67:969–75.
Oda Y, Wehrmann B, Radig K, Walter H, Röse I, Neumann W, et al. Expression of growth factors and their receptors in human osteosarcomas. Immunohistochemical detection of epidermal growth factor, platelet-derived growth factor and their receptors: its correlation with proliferating activities and p53 expression. Gen Diagn Pathol. 1995;141:97–103.
Sulzbacher I, Birner P, Trieb K, Träxler M, Lang S, Chott A. Expression of platelet-derived growth factor-AA is associated with tumor progression in osteosarcoma. Mod Pathol. 2003;16:66–71.
Hassan SE, Bekarev M, Kim MY, Lin J, Piperdi S, Gorlick R, et al. Cell surface receptor expression patterns in osteosarcoma. Cancer. 2012;118:740–9.
Andersen GB, Knudsen A, Hager H, Hansen LL, Tost J. miRNA profiling identifies deregulated miRNAs associated with osteosarcoma development and time to metastasis in two large cohorts. Mol Oncol. 2018;12:114–31.
Vimalraj S, Saravanan S, Raghunandhakumar S, Anuradha D. Melatonin regulates tumor angiogenesis via miR-424-5p/VEGFA signaling pathway in osteosarcoma. Life Sci. 2020;256:118011.
Wang J-C, Li G-Y, Wang B, Han S-X, Sun X, Jiang Y-N, et al. Metformin inhibits metastatic breast cancer progression and improves chemosensitivity by inducing vessel normalization via PDGF-B downregulation. J Exp Clin Cancer Res CR. 2019;38:235.
Cao R, Björndahl MA, Religa P, Clasper S, Garvin S, Galter D, et al. PDGF-BB induces intratumoral lymphangiogenesis and promotes lymphatic metastasis. Cancer Cell. 2004;6:333–45.
Thijssen VL, Paulis YW, Nowak-Sliwinska P, Deumelandt KL, Hosaka K, Soetekouw PM, et al. Targeting PDGF-mediated recruitment of pericytes blocks vascular mimicry and tumor growth. J Pathol. 2018;246:447–58.
Guo Y, Yin J, Wang Z, Zha L. Overexpression of platelet-derived growth factor-B increases the growth, invasion, and angiogenesis of gastric carcinoma cells through protein kinase B. Neoplasma. 2013;60:605–12.
Perut F, Roncuzzi L, Zini N, Massa A, Baldini N. Extracellular nanovesicles secreted by human osteosarcoma cells promote angiogenesis. Cancers. 2019;11:779.
Raica M, Cimpean AM. Platelet-derived growth factor (PDGF)/PDGF receptors (PDGFR) axis as target for antitumor and antiangiogenic therapy. Pharmaceuticals (Basel). 2010;3:572–99.
Wang L-H, Tsai H-C, Cheng Y-C, Lin C-Y, Huang Y-L, Tsai C-H, et al. CTGF promotes osteosarcoma angiogenesis by regulating miR-543/angiopoietin 2 signaling. Cancer Lett. 2017;391:28–37.
Lederle W, Stark H-J, Skobe M, Fusenig NE, Mueller MM. Platelet-derived growth factor-BB controls epithelial tumor phenotype by differential growth factor regulation in stromal cells. Am J Pathol. 2006;169:1767–83.
Lokker NA, Sullivan CM, Hollenbach SJ, Israel MA, Giese NA. Platelet-derived growth factor (PDGF) autocrine signaling regulates survival and mitogenic pathways in glioblastoma cells. Cancer Res. 2002;62:3729.
Furuhashi M, Sjöblom T, Abramsson A, Ellingsen J, Micke P, Li H, et al. Platelet-derived growth factor production by B16 melanoma cells leads to increased pericyte abundance in tumors and an associated increase in tumor growth rate. Cancer Res. 2004;64:2725–33.
Roberts WG, Whalen PM, Soderstrom E, Moraski G, Lyssikatos JP, Wang H-F, et al. Antiangiogenic and antitumor activity of a selective PDGFR tyrosine kinase inhibitor, CP-673,451. Cancer Res. 2005;65:957–66.
Sajib S, Zahra FT, Lionakis MS, German NA, Mikelis CM. Mechanisms of angiogenesis in microbe-regulated inflammatory and neoplastic conditions. Angiogenesis. 2018;21:1–14.
Vimalraj S, Saravanan S, Anuradha D, Chatterjee S. Models to investigate intussusceptive angiogenesis: a special note on CRISPR/Cas9 based system in zebrafish. Int J Biol Macromol. 2019;123:1229–40.
Plate KH, Scholz A, Dumont DJ. Tumor angiogenesis and anti-angiogenic therapy in malignant gliomas revisited. Acta Neuropathol. 2012;124:763–75.
Acknowledgements
This work was supported by Department of Science and Technology, INSPIRE Faculty Program, Government of India for the research grant to SV (grant no. DST/INSPIRE/04/2017/002913).
Author information
Authors and Affiliations
Contributions
SV, RS, SS, and BA performed all the experiments and drafted the manuscript. DA provided technical support for the work. SV secured the funding, designed the work, and analyzed data, and approved the final submitted manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Vimalraj, S., Subramanian, R., Saravanan, S. et al. MicroRNA-432-5p regulates sprouting and intussusceptive angiogenesis in osteosarcoma microenvironment by targeting PDGFB. Lab Invest 101, 1011–1025 (2021). https://doi.org/10.1038/s41374-021-00589-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-021-00589-3