Abstract
Accumulating evidences have suggested that extracellular vesicles (EVs) are crucial players in the pathogenesis of ischemic brain injury. This study was designed to explore the specific functions of M2 phenotype microglia-derived EVs in ischemic brain injury progression. The expression of microRNA-135a-5p (miR-135a-5p) in M2 microglia-derived EVs was determined by reverse transcription-quantitative polymerase chain reaction (RT-qPCR), followed by the identification of expression relationship among miR-135a-5p, thioredoxin-interacting protein (TXNIP), and nod-like receptor protein 3 (NLRP3) by dual luciferase reporter gene assay. After construction of an oxygen-glucose deprivation/reperfusion (OGD/R) cell model, the effects of miR-135a-5p on the biological characteristics of HT-22 cells were assessed by cell counting kit 8 (CCK-8) assay and flow cytometry. Finally, a mouse model of transient middle cerebral artery occlusion (tMCAO) was established and cerebral infarction volume was determined by triphenyltetrazolium chloride (TTC) staining and the expression of IL-18 and IL-1β in the brain tissue was determined by enzyme-linked immunosorbent assay (ELISA). We found that M2 microglia-derived EVs had high expression of miR-135a-5p, and that miR-135a-5p in M2 microglia-derived EVs negatively regulated the expression of NLRP3 via TXNIP. Overexpression of miR-135a-5p promoted the proliferation but inhibited the apoptosis of neuronal cells, and inhibited the expression of autophagy-related proteins. M2 microglia-derived EVs delivered miR-135a-5p into neuronal cells to inhibit TXNIP expression, which further inhibited the activation of NLRP3 inflammasome, thereby reducing neuronal autophagy and ischemic brain injury. Hence, M2 microglia-derived EVs are novel therapeutic targets for ischemic brain injury treatment.
Similar content being viewed by others
Introduction
Ischemic brain injury is induced by insufficient blood supply, which leads to energy depletion, loss of mitochondrial function, and nerve cell apoptosis [1, 2]. Brain ischemia is one of the most common causes of death and disability worldwide, thus posing an alarming health threat and adding a huge financial burden globally [3]. Microglial cells, as the primary immune cells in brain, are crucial mediators in the progression of brain ischemia [4, 5]. Extracellular vesicles (EVs), a family of membrane-bounded vesicles that are secreted from multiple cells including immune cells, can greatly affect intercellular communications through effectively delivering cargos of proteins, lipids, and various nucleic acids, and can be applied to treat many diseases [6]. EVs can be further divided into exosomes and microvesicles, which are all nano- to micrometer-sized vesicles [7]. Moreover, a previous study has confirmed that EVs derived from microglial cells are mediators of the progression of neuroinflammatory diseases [8]. Hence, the current study was designed to explore the underlying mechanism and specific functions of EVs derived from M2 microglial cells in ischemic brain injury.
MicroRNAs (miRNAs) are a group of small single-stranded non-coding RNAs, which are proposed to be important regulators in the occurrence and development of cerebral ischemia [9]. For instance, miR-135a-5p was confirmed to protect against the development of cerebral hypoxia/reoxygenation injury [10]. Also, miRNAs can be delivered by EVs to target cells and furthermore be used as potential biomarkers in various pathological conditions such as ischemia [11,12,13]. Thioredoxin-interacting protein (TXNIP) is an innate suppressor of thioredoxin [14]. Meanwhile, thioredoxin is a small molecular weight redox protein that participates in maintaining the redox balance in living cells [15]. Previous research revealed that lowered expression of TXNIP in neurons can bring about an amelioration of ischemic brain injury [16]. Nod-like receptor protein 3 (NLRP3) inflammasome, which is a member of NOD-like receptor family, plays a central role in inflammatory responses that are associated with diverse inflammatory diseases [17]. Previous work has demonstrated that NLRP3 downregulation can alleviate aspects of the pathology due to cerebral ischemia [18]. Furthermore, the NLRP3 inflammasome was observed to be regulated by TXNIP, and is thus implicated in the regulation of damage following cerebral artery occlusion [19]. In addition, TXNIP-mediated NLRP3 expression can be reduced by miR-17-5p overexpression to serve as therapeutic pathway for neonatal hypoxic-ischemic brain injury in rats [20]. Based on the aforementioned findings, we hypothesized that EVs derived from M2 microglial cells were involved in the development of ischemic brain injury through TXNIP-mediated NLRP3 repression via miR-135a-5p.
Materials and methods
Bioinformatic analysis
Key miRNAs for ischemic brain injury were obtained through reviewing existing literature, and the downstream genes of miRNAs were predicted using the databases TargetScan (Total context + + score < −0.3) (http://www.targetscan.org/vert_72/), RAID (Score > 0.7) (http://www.rna-society.org/raid2/index.html), mirDIP (Integrated Score > 0.65, Number of Sources > 15) (http://ophid.utoronto.ca/mirDIP/), RNA22 (https://cm.jefferson.edu/rna22/) and microRNA (energy < −15, mirsvr_score < −0.55) (http://www.microrna.org/microrna/home.do), respectively. Then. an intersection was taken by plotting the Venn diagram among the predicted genes. String (minimum required interaction score: 0.150) (https://string-db.org) was employed to predict the relevant genes obtained from the intersection and to construct a protein–protein interactions (PPI) network. Cytoscape (https://cytoscape.org) was used to draw diagrams of the network and calculate the core degree of the genes; the gene with the highest core degree in the brain injury network was selected as the key gene, and its possible downstream pathway was obtained by combining data from the existing studies. The binding site of miRNA and gene was predicted through TargetScan and the co-expression of key genes and their downstream pathways was detected through Multi Experiment Matrix (MEM) (https://biit.cs.ut.ee/mem/index.cgi).
Activation and identification of M2 microglial cells
BV2 cells purchased from the Chinese Academy of Sciences Cell Bank were cultured in Dulbecco’s modified minimal medium (DMEM, HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS, GIBCO BRL, Grand Island, NY, USA) and 1% penicillin-streptomycin antibiotic (HyClone). Next, the BV2 cells were treated with interleukin 4 (IL-4, 20 ng/mL, HEILP-0401, Cyagen, Santa Clara, CA, USA) for 48 h to activate M2 phenotype polarization followed by identification with immunocytochemistry (ICC) and polymerase chain reaction (PCR).
Oxygen-glucose deprivation/reperfusion (OGD/R) cell model construction
Mouse hippocampal neuronal cells HT-22 purchased from the Chinese Academy of Sciences Cell Bank were cultured in a humidified incubator at 37 °C in 5% CO2 with DMEM medium (BasalMedia, Shanghai, China) containing 10% FBS. The medium was renewed every 2 days. Cell passage was conducted when the cell confluence reached 90%. After being cultured for 7–10 days, HT-22 cells were washed twice with DMEM without glucose (BasalMedia, Shanghai, China), and incubated in deoxygenated glucose-free DMEM (BasalMedia, Shanghai, China) containing 10% FBS in a hypoxic incubator (95% N2 and 5% CO2) to conduct OGD experiments. After 45 min, cells were removed from the incubator, the OGD solution in the culture was replaced with maintenance medium, and then the cells were placed for recovery in a conventional incubator for 12 h and then collected for subsequent analysis.
Lentivirus infection
The TXNIP lentiviral overexpression vector purchased from Genechem (Shanghai, China) was cloned into a self-inactivating lentiviral vector (pSicoR). The synthesized oligonucleotides were annealed and inserted into the cloning restriction sites of Hpa 1 and Xho I, positive clones were identified by restriction enzyme digestion and DNA sequencing, and then Lipofectamine 2000TM (Invitrogen, Carlsbad, CA, USA) was used to transfect lentiviral vectors and packaging vectors (psPAX2 and pMD2.G) containing positive inserts into 293 T cells. From 24 to 72 h after transfection, the supernatant containing lentiviral particles was collected every 12 h and filtered through a 0.45 μm cellulose acetate filter and formulated to give a final virus titer of 109 TU/mL. One day before infection, neuronal cells were detached by 0.25% trypsin, modeled as above, and counted. Cells were then seeded into a six-well plate at a cell density of 2 × 105 cells/well, and cultured in a DMEM medium with 10% FBS containing EVs (A2720801, GIBCO BRL, Grand Island, NY, USA) and placed in the incubator at 37 °C with 5% CO2. When the cell confluence reached 30–50%, cells were added with 1 mL/volume of complete culture solution already containing tenfold dilution of the virus (the dilution ratio at 103–107) and Polybrene (H8761, Solarbio, Beijing, China) for incubation at 37 °C with 5% CO2. On the second day, the virus-containing culture medium was aspirated, and 2 mL of fresh complete culture medium was added for culturing overnight. After 5 days of culturing, the GFP expression in cells was measured under a fluorescent microscope, which showed a > 95% rate of GFP positive cells. After infection with lentiviral particles, cells stably transfected with lentivirus were screened in medium containing 0.5 μg/mL puromycin.
Cell transfection
M2-BV2 cells were transfected with miR-135a-5p mimic, miR-135a-5p inhibitor, and their negative controls (NC mimic, NC inhibitor). The modeled HT-22 cells were transfected with miR-135a-5p mimic, miR-135a-5p inhibitor, miR-135a-5p mimic + TXNIP and their negative controls (NC mimic, NC inhibitor, miR-135a-5p mimic + NC mimic).
One day before transfection, M2-BV2 cells and HT-22 cells in the logarithmic growth phase were inoculated into a 6-well plate at a concentration of 1 × 105 cells/mL. When cell confluence reached 50–70%, 800 μL of serum-free medium was added to each well, and then the mixture of the above mimic, inhibitor or plasmid (all purchased from Biomics Biotech, Nantong, Jiangsu, China) and lipo2000 (11668027, Thermo Fisher Scientific, Waltham, MA, USA) was added to the six-well plate. After 6 h in culture, the medium was renewed, and after 24 h of transfection, cells were screened with the antibiotic G418 at a final concentration of 600 mg/L, and the medium was changed every 3 days. After 12 days in culture, the M2-BV2 cells and HT-22 cells that had received no transfection all died while 48% of the cells receiving transfection were still alive. Afterward, the culture was further performed at a maintenance concentration of 300 mg/L G418 to obtain stable expression cell lines.
Isolation of EVs
M2-BV2 cells at logarithmic growth period were cultured at 37 °C with 5% CO2 using DMEM medium without EVs (10% FBS and 1% penicillin-streptomycin antibiotics), the cell supernatant was collected 2 days after culture, followed by filtration using a 0.22 μm filter to remove cell clumps. After gradient centrifugation, the liquid at the bottom of the centrifuge tube were retained and the supernatant discarded. Then cells were resuspended in sterile PBS, centrifuged at 110000 r/min at 4 °C for 90 min to obtain a precipitate, which was then resuspended in 10 mL of phosphate buffer (PBS). The left pellets after centrifugation at 4 °C and 110,000 r/min for 90 min were designated as EVs, which were resuspended in 100 μL PBS and stored at −80 °C until use.
Transmission electron microscope (TEM) observation
EVs were precipitated and then immediately fixed in 2.5% glutaraldehyde at 4 °C. After fixation, the specimens were dehydrated with a gradient alcohol and immersed in epoxy resin for embedding. The ultrathin sections were stained with uranyl acetate and lead citrate and observed under a TEM (JEM-1010, JEOL, Tokyo, Japan).
Nanoparticle tracking analysis (NTA)
The collected EVs were diluted with PBS to a particle concentration of 106 cells/mL-109 cells/mL, and then a 1 mL syringe was used to draw the sample into a Nanosight analyzer (Nanosight NS300, Malvern Panalytical, Worcestershire, UK) for detection and analysis.
EV internalization
EVs were incubated with 2 μM red fluorescent dye PKH26 (Sigma-Aldrich, St. Louis, MO, USA) for 5 min and washed 5 times through a 100 kDa filter (Microcon YM-100) to remove excess staining solution. HT-22 cells after modeling were plated in a six-well plate of neuronal basal medium at a density of 5 × 105 cells/well. The labeled EVs extracted from M2-BV2 cells were co-cultured with neuronal cells at the concentration of 100 μg/mL in an incubator with 5% CO2 at 37 °C and saturated humidity. Neuronal cells were cultured in serum for 12 h before the stimulation experiment. After incubation for 3 days, images were taken with a Zeiss LSM 780 confocal microscope (Zeiss, Jena, Germany).
Cell counting kit 8 (CCK-8) assay
CCK-8 kit (WH1199, Biotechwell Co., Ltd., Shanghai, China) was used to detect cell proliferation. Cells at logarithmic growth phase were adjusted to 5 × 104 cells/mL using DMEM medium containing 10% FBS, and then seeded to a 96-well culture plate at 100 μL/well and cultured. Next, 10 μL CCK-8 solution was added into each well at the corresponding time points, and then the optical density (OD) value at wavelength of 450 nm was measured by Multiskan FC microplate reader (51119080, Thermo Fisher Scientific) after incubation at 37 °C for 2 h. Five parallel wells were set and the average value was taken.
Flow cytometry
HT-22 cells were seeded in a 24-well plate. After 24 h of culturing, cells were detached with trypsin. After 24 h, cells were mixed gently with 400 μL containing Annexin-V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) binding buffer (PromoKine, Heidelberg, Germany), incubated at room temperature for 10 min in the dark. Flow cytometer (Becton Dickinson, USA) was excited at a wavelength of 488 nm, AnnexinV at a wavelength of 525/530 nm, PI at a wavelength of 690/50 nm, and cell apoptosis was detected using Guava Easy Cyte 6-2 L system (Merck Millipore, Billerica, MA, USA).
Transient middle cerebral artery occlusion (tMCAO) mouse model construction
Sixty adult male ICR mice weighing 25–30 g (provided by Shanghai Laboratory Animal Center, Chinese Academy of Sciences) were fed with standard mouse chow with free access to water and a light/dark cycle of 12 h/12 h. Mice were anesthetized with 1.5–2% isoflurane and 30%/70% oxygen/nitrous oxide, and a silicone-coated 6-0 nylon suture was inserted through the external carotid artery, the internal carotid artery, and then the middle cerebral artery of the mice. An occlusion was considered successful a when the cortical surface cerebral blood flow (CBF) lowered to 10% of the baseline value, as measured using a laser Doppler flowmeter (Moor Instruments, Devon, UK). After the occlusion for one hour, the suture was removed for reperfusion. Immediately after suture removal, EVs were immediately injected into the mice through a tail vein. The injection was repeated for 3 days at a dose of 100 μg/kg per day.
Brain weight measurements
After treatment with EVs, tMCAO mice were euthanized under deep anesthesia, and the brain was removed without pre-perfusion. The cerebellum and brain stem were dissected from the forebrain, and the cerebral hemispheres were separated along the mid-line and weighed with a high-precision balance (sensitivity 1 mg). The results were expressed as the mass ratio of the ipsilateral (right) and contralateral (left) hemispheres of mice.
Triphenyltetrazolium chloride (TTC)
After treatment with EVs, tMCAO mice were euthanized under deep anesthesia, and the brain was removed as above and cut into 2 mm sections with a vibrotome, and the brain sections were incubated in the dark in 1% tetracycline (TTC) solution (SR0229, Oxiod, Thermo Fisher Scientific) for 5 min and then fixed with 10% formaldehyde and mounted for microscopic examination. Image J software (Version 1.43 u, National Institutes of Health, Bethesda, MD, USA) was used to track and analyze the volume of cerebral infarction. The following formula was used to calculate the percentage of infarction: (total area of contralateral hemisphere-uninfarcted area of ipsilateral hemisphere)/(total area of contralateral hemisphere × 2).
Enzyme-linked immunosorbent assay (ELISA)
After treatment with EVs, tMCAO mice were euthanized under deep anesthesia, and the brain was collected without pre-perfusion. The mouse brain was placed in a 2.0 centrifuge tube, and centrifuged at 3000 r/min for 10 min at 4 °C with the supernatant collected. The concentration of IL-18 and IL-1β in the supernatant was detected by an ELISA kit (69-21183; 69-21178, MSKBio Co., Ltd., Wuhan, Hubei, China) in strict accordance with the standard procedures on the instructions. The OD value of each well at 450 nm was measured using a universal microplate reader (Synergy 2, BioTek, Biotek Winooski, Vermont, USA). Taking the standard concentration as the abscissa and the OD value as the ordinate, the regression equation of the standard curve was calculated, and the OD value of sample was substituted into the equation to calculate the target protein concentration in the sample.
Dual-luciferase reporter gene assay
HEK-293T cell line (purchased from the Cell Bank of Shanghai Institute of Cytology, Chinese Academy of Sciences) was cultured in DMEM medium. When the cell density reached 80–90%, cells were detached with 0.25% trypsin, passaged, and cultured in a constant temperature incubator with 5% CO2 at 37 °C. Cells in logarithmic growth phase were taken for experiments. The biological prediction website Targetscan.org was used to analyze the target gene of miR-135a-5p, and then the dual-luciferase reporter gene assay was used to verify whether TXNIP was the direct target gene of miR-135a-5p. The synthetic TXNIP 3′UTR gene fragment was introduced into the pmirGLO vector (Promega, Madison, WI, USA) using the endonuclease site, and the complementary sequence mutation (MUT) site of the seed sequence was designed on the TXNIP wild type (WT) through the use of restriction enzyme. The target fragment was inserted into the pGL3-control vector using T4 DNA ligase. After confirming the sequence, the luciferase reporter plasmids WT and MUT were co-transfected with miR-135a-5p mimic into HEK-293T cells. Cells were collected and lysed 48 h after transfection, and the luciferase activity was detected on a Luminometer TD-20/20 detector (E5311, Promega) using the Dual-Luciferase Reporter Assay System kit (Promega).
miRNA analysis
miRNAs in EVs were isolated using SeraMir exosome RNA Purification Kit (System Biosciences, Mountain View, CA, USA), and total RNA was extracted from cultured cells using Trizol reagent (15596026, Invitrogen). Subsequently, miRNA was reversely transcribed according to the instructions of the miScript Reverse Transcription Kit (Qiagen GmbH, Hilden, Germany). The primer sequences of miR-135a-5p are F: 5′-CCGGCGTATGGCTTTTTATTCC-3′; R: 5′-CAGTGCAGGGTCCGAGGT-3′, and U6 (F: 5′-CTCGCTTCGGCAGCACATATACT-3′; R: 5′-ACGCTTCACGAATTTGCGTGTC-3′) was taken as internal control. Quantitative PCR detection was performed according to the instructions of Stem-Loop Detection Kit (GenePharma, Shanghai, China). The relative transcription level of the target gene mRNA was calculated by the 2-ΔΔCt method. Each sample was analyzed in triplicate and the average value was taken.
Reverse transcription-quantitative PCR (RT-qPCR)
PrimeScriptTM RT Reagent Kit (Perfect Real Time) (Takara, Japan) was used to synthesize 1 μg of total RNA into cDNA. Then, the StepOnePlusTM Real-Time PCR System (Invitrogen) was used to perform RT-qPCR analysis of TXNIP, Arginase 1, IL-10, and CD206. The primer sequences are presented in Table 1. Using β-actin as an internal reference, SYBR® Premix Ex TaqTM (Tli RNaseH Plus, Takara) was used for amplification. The relative transcription level of the target gene mRNA was taken as the CT value (inflection point of the amplification power curve). The 2-ΔΔCt method was used to indicate relative expression of each gene.
Western blot analysis
Cells or tissues were added with 1 mL of cell lysis buffer (containing protease inhibitors) (P0013J, Beyotime, Shanghai, China) for 45 min and lysed for 30 min at 4 °C and 8000 rpm. The supernatant was collected and the protein concentration of each sample was determined using a bicinchoninic acid (BCA) kit (PC0020, Solarbio). Then, 50 μg protein was taken from each group for electrophoretic separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then the proteins were transferred onto a polyvinylidene fluoride membrane (66485, PALL, USA) by the wet method. The membrane was blocked with 5% skimmed milk powder, sealed at room temperature for 2 h, washed with TBST three times, and incubated with primary antibodies: TXNIP (# 14715, 1: 1000, CST, Danvers, MA, USA), Beclin-1 (ab207612, 1: 2000, Abcam, Cambridge, UK), light chain3A/B (LC3A/B; #12741, 1: 1000, CST), lysosomal-associated membrane protein 1 (LAMP-1; ab108597, 1: 1000, Abcam), P62 (ab155686, 1: 3000, Abcam), nod-like receptor protein 3 (NLRP3; #15101, 1: 1000, CST), pro-IL-1β (ab2105, 1: 1000, Abcam), Caspase-1 p20 (22915-1-AP, 1: 2000, Proteintech, Rosemont, IL, USA), tumor susceptibility gene 101 (TSG101; ab125011, 1: 1000, Abcam), CD63 (25682-1-AP, 1: 1000, Proteintech), heat shock protein 70 (Hsp70; ab181606, 1: 1000, Abcam), calnexin (ab10286, 1: 2000, Abcam), and β-actin (ab8227, 1: 1000, Abcam) at 4 °C overnight. Then the membrane was added with horseradish peroxidase (HRP)-labeled immunoglobulin G (IgG, ab6721, 1: 2000, Abcam) secondary antibody working solution of the corresponding species and incubated at room temperature for 1 h followed by immersion in enhanced chemiluminescence (ECL) reaction solution (BM101, Biomiga, San Diego, CA, USA) at room temperature for 1 min. Observation was conducted with a chemiluminescence instrument in the dark. β-actin was taken as internal reference, and the ratio of the gray value of the target band to the internal reference band was used as the relative expression of the protein.
Statistical analysis
The SPSS 21.0 statistical software (IBM Corp, Armonk, NY, USA) was used for statistical analysis. Measurement data were expressed as mean ± standard derivation. Comparison between groups was analyzed by independent t-test. Comparisons among multiple groups were analyzed by one-way analysis of variance (ANOVA) and followed by Tukey’s post hoc test. A value of p < 0.05 was considered statistically significant.
Results
miR-135a-5p modulates ischemic brain injury predicted by bioinformatics analysis
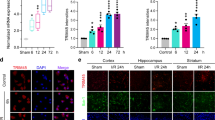
It has been reported that EVs derived from M2 microglia can protect the brain of mice from ischemia-reperfusion injury [21]. In addition, miR-135a is a regulator of central nervous system growth and migration in vivo and in vitro, and can reduce neuronal damage induced by OGD/R [22]. Thus, we tried to focus on the regulatory role of miR-135a-5p in ischemic brain injury. First, the downstream genes of miR-135a-5p were predicted using the TargetScan, RAID, mirDIP, RNA22, and microRNA databases, which obtained 312, 34, 421, 10905, and 358 genes, respectively. Three key downstream genes NR3C2, VLDLR, and TXNIP were obtained from the intersection (Fig. 1A). String was used to predict the related genes of these three important genes and a PPI network was established. The core level was mapped and calculated by Cytoscape, which showed that the VLDLR and TXNIP genes both had a core level of 5, whereas the core degree of NR3C2 was 2 (Fig. 1B). TXNIP has been demonstrated to aggravate brain injury by activating NLRP3 inflammasomes, and this relationship was also been predicted in the PPI network. The binding site map of miR-135a-5p and TXNIP was predicted by TargetScan (Fig. 1C). The co-expression relationship between TXNIP and NLRP3 was explored by MEM analysis, the results of which indicated an obvious co-expression between these two genes (Fig. 1D). Hence, TXNIP and NLRP3 were identified as two downstream genes of miR-135a and their specific functions were investigated subsequently.
A Venn diagram of the prediction results of the downstream genes of miR-135a-5p from the TargetScan, RAID, mirDIP, RNA22, and microRNA databases. B PPI network of 3 intersection genes constructed by String; squares are input genes, circles are predicted relevant genes, deeper red indicates the genes with higher the core degree, and blue a lower the core degree. C The binding sites of miR-135a-5p and TXNIP in human (top) and mouse (bottom) predicted by TargetScan. D The co-expression relationship between TXNIP and NLRP3 predicted by MEM analysis.
EVs derived from M2 microglia inhibits neuronal autophagy and ischemic brain injury
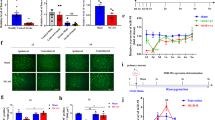
In order to study the mechanism of M2 microglia-derived EVs in ischemic brain injury, M2 polarization was primarily induced in BV2 cells. The results of ICC revealed that, compared with that of BV2 cells, the expression of microglia activation indicator CD68 in M2-BV2 cells increased notably (Fig. 2A). RT-qPCR demonstrated that, compared with that of BV2 cells, the expression of M2 microglia markers Arginase 1, IL-10, and CD206 in M2-BV2 cells were increased (Fig. 2B). M2-BV2 cells were selected and then EVs produced by M2-BV2 cells were separated and purified. A group of membranous vesicles with obvious heterogeneity in size and round or elliptical membranes were observed under TEM, in which the membranous structure could be seen on the periphery of the vesicle, with a low electron density component in the center (Fig. 2C). According to the results of the Nanosight Nanoparticle Tracking Analyzer, EVs exhibited irregular Brownian motion indicating a diameter between 30 and 120 nm (Fig. 2D). Results of Western blot analysis proved that EVs expressed EV marker proteins TSG101, CD63, and Hsp70, but did not express calnexin (Fig. 2E).
A The expression of microglia activation markers determined by ICC. B The expression of M2 microglia markers determined by RT-qPCR. C Morphological characteristics of EVs observed by TEM (Scale bar = 100 nm). D EVs observed by Nanosight nanoparticle tracking analysis. E The expression of EV-related proteins determined by Western blot analysis. F The binding of fluorescence-labeled EVs and HT-22 cells received OGD/R modeling observed by laser confocal. G The expression of autophagy-related proteins measured by Western blot analysis. H The proliferation of HT-22 cells after OGD/R modeling and co-culture treatment detected by CCK-8. I The apoptosis of HT-22 cells after OGD/R modeling and co-culture treatment detected by flow cytometry. J Brain mass ratio of the ipsilateral (right) and contralateral (left) hemispheres of mice after tMCAO modeling (n = 6). K Cerebral infarction in mice after tMCAO modeling detected by TTC (n = 6). L The concentration of IL-18 and IL-1β in the brain tissue supernatant of mice after tMCAO modeling determined by ELISA (n = 6). Comparison between two groups was analyzed by independent t test, and comparison among multiple groups was analyzed by one-way ANOVA and followed by Tukey’s post hoc test. *p < 0.05. Cell experiments were repeated three times.
In order to verify whether EVs derived from M2 microglia can enter neuronal cells and affect their function, an OGD/R model was established in HT-22 cells, which were then incubated with the PKH26 labeled EVs. After 6 h of culture, PKH26 red fluorescence was observed in HT-22 cells, indicating that PKH26-labeled EVs were internalized by HT-22 cells (Fig. 2F). Then the effect of M2 microglia-derived EVs on neuronal cells was detected through a series of experiments. We found that, compared with cells after sham operation, the expression of P62 (Fig. 2G) in HT-22 cells and proliferation in cells (Fig. 2H) after OGD/R modeling were both reduced remarkably, whereas the expression of Beclin-1, LC3A/B, and LAMP-1 (Fig. 2G), and cell apoptosis (Fig. 2I) were increased. Compared with cells treated by OGD/R + PBS, with increasing concentration of EVs (50 μg/mL in the L-EVs group, 70 μg/mL in the M-EVs group was, and 100 μg/mL in the H-EVs group), the expression of P62 (Fig. 2G) and cell proliferation (Fig. 2H) increased, while the expression of Beclin-1, LC3A/B and LAMP-1 (Fig. 2G) and cell apoptosis (Fig. 2I) were both lower in HT-22 cells treated with OGD/R.
Subsequently, the tMCAO model was constructed in mice followed by intravenous treatment with EVs. We observed that, compared with sham-operated mice, the mass ratio of the brain on the ipsilateral (right) and contralateral (left) hemispheres (Fig. 2J) of tMCAO model mice was lowered, but the cerebral infarction rate (Fig. 2K) and the concentration of IL-18 and IL-1β (Fig. 2L) in the brain supernatant were increased markedly. Compared with mice treated by tMCAO + PBS, with increasing concentration of EVs, the mass ratio of the ipsilateral (right) and contralateral (left) hemispheres (Fig. 2J) of tMCAO mice showed an increasing trend, and the cerebral infarction rate (Fig. 2K) and the concentrations of IL-18 and IL-1β in the brain tissue supernatant both decreased (Fig. 2L). In summary, M2 microglia-derived EVs inhibited the autophagic activity of neurons, while promoting their proliferation, and thus repressing ischemic brain injury.
EVs derived from M2 microglia inhibits neuronal autophagy and ischemic brain injury by delivering miR-135a-5p
In order to further study the mechanism of M2 microglia-derived EVs affecting neuronal cells, M2-BV2 cells were transfected with miR-135a-5p mimic, miR-135a-5p inhibitor, and their negative controls (NC mimic, NC inhibitor), respectively, and changes in the miR-135a-5p expression in the M2-BV2 cells were determined by RT-qPCR. At 48 h after transfection, the expression of miR-135a-5p in MV-BV2 cells transfected with miR-135a-5p mimic was increased, and the expression of miR-135a-5p in M2-BV2 cells transfected with miR-135a-5p inhibitor was reduced (Fig. 3A). Then RT-qPCR results illustrated that, compared with M2-BV2 cells treated by EVs-NC mimic, the expression of miR-135a-5p in cells treated by EVs-miR-135a-5p mimic was increased. Compared with EVs-NC inhibitor treatment, expression of miR-135a-5p after EVs-miR-135a-5p inhibitor treatment was reduced (Fig. 3B).
A The transfection efficiency of miR-135a-5p detected by RT-qPCR. B The expression of miR-135a-5p in the EVs derived from M2-BV2 cells measured by RT-qPCR. C The expression of miR-135a-5p in HT-22 cells after transfection measured by RT-qPCR. D The expression of autophagy-related proteins in HT-22 cells after transfection measured by Western blot analysis. E The proliferation of HT-22 cells after OGD/R treatment and transfection detected by CCK-8 assay. F The apoptosis of HT-22 cells after OGD/R treatment and transfection detected by flow cytometry. Comparison between two groups was analyzed by independent t-test, and comparison among multiple groups was analyzed by one-way ANOVA and followed by Tukey’s post hoc test. *p < 0.05. Cell experiments were repeated three times.
Then, the EVs were cultured with HT-22 cell models. Compared with HT-22 cells co-cultured with EVs-NC mimic, the expression of miR-135a-5p in HT-22 cells co-cultured with EVs-miR-135a-5p mimic elevated. Compared with EVs-NC inhibitor, the expression of miR-135a in HT-22 cells cultured with EVs-miR-135a-5p inhibitor decreased (Fig. 3C).
Then a series of experiments were conducted to study the effect of M2 microglia-derived EVs on neuronal cells, with results showing that, compared with HT-22 cells co-cultured with EVs-NC mimic, the expression of P62 (Fig. 3D) and cell proliferation (Fig. 3E) were both promoted, while the expression of Beclin-1, LC3A/B and LAMP-1 (Fig. 3D) and cell apoptosis (Fig. 3F) were both inhibited in HT-22 cells co-cultured with EVs-miR-135a-5p mimic. Opposite effects were seen in HT-22 cells co-cultured with EVs-miR-135a-5p inhibitor compared with HT-22 cells co-cultured with EVs-NC inhibitor (Fig. 3D–F). To sum up, M2 microglia-derived EVs containing miR-135a-5p inhibited the autophagic activity of neurons, promoted cell proliferation, and thus reducing ischemic brain injury.
M2 microglia-derived EVs inhibit TXNIP expression by delivering miR-135a-5p
Through prediction results of online websites, there was specific binding between miR-135a-5p and TXNIP. Next, dual-luciferase reporter gene assay was used to verify that miR-135a-5p could bind to TXNIP. Results showed, that compared with NC mimic + TXNIP-3′UTR-WT, the fluorescence intensity of cells treated by miR-135a-5p mimic + TXNIP-3′UTR-WT decreased. Compared with cells treated by NC mimic + TXNIP-3’UTR-MUT, there was no significant change in fluorescence intensity in cells treated by miR-135a-5p mimic + TXNIP-3′UTR-MUT (Fig. 4A), indicating that miR-135a-5p can specifically bind to TXNIP.
A,The binding between miR-135a-5p and TXNIP explored by dual luciferase reporter gene assay. B The expression of TXNIP in HT-22 cells after modeling measured by RT-qPCR. C The expression of miR-135a-5p in HT-22 cells after modeling and transfection measured by RT-qPCR. D The expression of TXNIP in HT-22 cells after modeling measured by Western blot analysis. E The expression of TXNIP in modeled HT-22 cells with EV treatment measured by Western blot analysis. F The expression of TXNIP in modeled HT-22 cells measured by RT-qPCR. G The expression of miR-135a-5p and TXNIP in modeled HT-22 cells without and with EV treatment measured by RT-qPCR. H The expression of autophagy-related proteins modeled in HT-22 cells without and with EVs treatment determined by Western blot analysis. I The proliferation of modeled HT-22 cells without and with EV treatment detected by CCK-8. J The apoptosis of modeled HT-22 cells without and with EV treatment detected by flow cytometry. Comparison between two groups was analyzed by independent t-test, and comparison among multiple groups was analyzed by one-way ANOVA and followed by Tukey’s post hoc test. *p < 0.05. Cell experiment was repeated three times.
Then OGD/R model was constructed in HT-22 cells, and the expression of TXNIP after modeling was determined by RT-qPCR. Compared with cells after the sham procedure, the expression of TXNIP in HT-22 cells after OGD/R modeling was up-regulated (Fig. 4B). To verify whether miR-135a-5p can regulate the expression of TXNIP in neuronal cells, cells were transfected with miR-135a-5p mimic, miR-135a-5p inhibitor, and their negative controls (NC mimic and NC inhibitor), and transfection efficiency was determined by RT-qPCR. At 24 h after transfection, the expression of miR-135a-5p in HT-22 cells transfected with miR-135a-5p mimic was elevated, and the expression of miR-135a-5p in HT-22 cells transfected with miR-135a-5p inhibitor down-regulated (Fig. 4C). Western blot analysis presented that, compared with NC mimic treatment, the expression of TXNIP in HT-22 cells transfected with miR-135a-5p mimic reduced. Compared with NC inhibitor, expression of TXNIP in HT-22 cells transfected with miR-135a-5p inhibitor was promoted (Fig. 4D).
In addition, EVs were extracted from M2-BV2 cells after transfection of miR-135a-5p mimic, miR-135a-5p inhibitor and their negative controls (NC mimic and NC inhibitor) and co-cultured with HT-22 cells after OGD/R modeling. Western blot analysis was used to measure the expression of TXNIP. Results revealed that, compared with HT-22 cells co-cultured with EVs-NC mimic, the expression of TXNIP in HT-22 cells co-cultured with EVs-miR-135a-5p mimic was lowered. Compared with HT-22 cells co-cultured with EVs-NC inhibitor, expression of TXNIP in HT-22 cells co-cultured with EVs-miR-135a-5p inhibitor elevated (Fig. 4E). To sum up, M2 microglia-derived EVs could deliver miR-135a-5p to inhibit TXNIP.
In order to verify whether the EVs derived from M2 microglia can inhibit TXNIP through the delivery of miR-135a-5p and thereby affect the function of neurons, cells were treated by overexpressed TXNIP and the transfection efficiency was detected by RT-qPCR. After 24 h, the expression of TXNIP in HT-22 cells after TXNIP treatment increased notably (Fig. 4F). Then, the EVs isolated from M2-BV2 cells overexpressing miR-135a-5p were co-cultured with the TXNIP overexpression-treated cells. Compared with the NC treatment, the expression of miR-135a-5p in HT-22 cells after TXNIP treatment was unaltered, but the expression of TXNIP was up-regulated. Compared with cells treated by TXNIP + EVs-NC mimic, the expression of miR-135a-5p in HT-22 cells treated with TXNIP + EVs-miR-135a-5p mimic was obviously increased, and the expression of TXNIP was remarkably reduced (Fig. 4G).
Then a series of experiments were employed to study the effect of M2 microglia-derived EVs on neuronal cells. Compared with NC treatment, the expression of P62 (Fig. 4H) and cell proliferative capacity (Fig. 4I) were suppressed, and the expression of Beclin-1, LC3A/B, and LAMP-1 (Fig. 4H) and cell apoptotic capacity (Fig. 4J) were enhanced in HT-22 cells after TXNIP treatment. Compared with cells treated by TXNIP + EVs-NC mimic, the expression of P62 (Fig. 4H) and cell proliferative capacity (Fig. 4I) were promoted, and the expression of Beclin-1, LC3A/B and LAMP-1 (Fig. 4H) and cell apoptotic capacity (Fig. 4J) were decreased in HT-22 cells treated by TXNIP + EVs-miR-135a-5p mimic (Fig. 4H–J). To sum up, the miR-135a-5p delivered by EVs derived from M2 microglia can repress TXNIP, thereby inhibiting the autophagic activity of neurons and promoting their proliferation, ultimately alleviating ischemic brain injury.
M2 microglia-derived EVs containing miR-135a-5p alleviates ischemic brain injury via TXNIP/NLRP3 axis
The expression of NLRP3 inflammasome in the cells after OGD/R modeling was determined by Western blot analysis. Compared with the cells after sham operation, the expression of NLRP3, pro-IL-1β, and Caspase-1 p20 increased in HT-22 cells after OGD/R modeling (Fig. 5A). Then cells after modeling were treated with TXNIP or miR-135a-3p mimic + TXNIP, and western blot analysis was conducted to measure the expression of NLRP3 inflammasome. Compared with cells transfected with NC, the expressions of NLRP3, pro-IL-1β, and Caspase-1 p20 in HT-22 cells treated by TXNIP were up-regulated. Compared with cells transfected with TXNIP + NC mimic, expressions of NLRP3, pro-IL-1β and Caspase-1 p20 in HT-22 cells treated by TXNIP + miR-135a-5p mimic were reduced (Fig. 5B).
A The expression of NLRP3 inflammasome in HT-22 cells after modeling measured by Western blot analysis. B The expression of NLRP3 inflammasome in HT-22 cells after modeling and transfection measured by Western blot analysis. C The expression of miR-135a-5p in modeled HT-22 cells with EV treatment measured by RT-qPCR. D The expression of TXNIP, autophagy-related protein and NLRP3 inflammasome in modeled HT-22 cells with EVs treatment measured by Western blot analysis. E The proliferation of HT-22 cells detected by CCK-8. F The apoptosis of HT-22 cells detected by flow cytometry. G The brain mass of mice after tMCAO modeling (n = 6). H The cerebral infarction in mice after tMCAO modeling determined by TTC (n = 6). I The concentration of IL-1β, IL-18 in brain supernatant of mice tissues after tMCAO modeling determined by ELISA (n = 6). Comparison between two groups was analyzed by independent t test, and comparison among multiple groups was analyzed by one-way ANOVA and followed by Tukey’s post hoc test. *p < 0.05. Cell experiments were repeated three times.
In order to further study the mechanism of M2 microglia-derived EVs on ischemic brain injury, EVs in M2-BV2 cells transfected with miR-135a-5p mimic were extracted and then cultured with HT-22 cells after OGD/R modeling or/and TXNIP treatment. Compared with EVs-NC mimic, the expression of miR-135a-5p in HT-22 cells treated by EVs-miR-135a-5p mimic was elevated (Fig. 5C). The expression of TXNIP, autophagy-related protein, and NLRP3 inflammasome-related detection indicators in the cells after modeling was measured by Western blot analysis. Compared with EVs-NC mimic, the expressions of TXNIP, Beclin-1, LC3A/B, LAMP-1, NLRP3, pro-IL-1β, and Caspase-1 p20 declined in HT-22 cells co-cultured with EVs-miR-135a-5p mimic, and the expression of P62 was up-regulated. Compared with EVs-miR-135a-5p mimic + NC treatment, the expressions of TXNIP, Beclin-1, LC3A/B, LAMP-1, NLRP3, pro-IL-1β, and Caspase-1 p20 was elevated in HT-22 cells co-cultured with EVs-miR-135a-5p mimic + TXNIP, and the expression of P62 was decreased (Fig. 5D).
The effect of M2 microglia-derived EVs on neuronal cells was then investigated through a series of experiments. Our findings uncovered that, compared with EVs-NC mimic, the proliferative ability was enhanced while the apoptosis ability was repressed in cells co-cultured with EVs-miR-135a-5p mimic. Compared with cells co-cultured with EVs-miR-135a-5p mimic + NC, the proliferative ability was repressed in HT-22 cells co-cultured with EVs-miR-135a-5p mimic + TXNIP, but the apoptotic ability was promoted (Fig. 5E, F).
Finally, EVs extracted from untreated M2-BV2 cells, those treated with miR-135a-5p, miR-135a-5p mimic + NC mimic, or miR-135a-5p mimic + TXNIP were injected into tMCAO mice by tail vein injection. Compared with EVs-NC mimic treatment, the mass ratio of the ipsilateral (right) and contralateral (left) hemispheres (Fig. 5G) of mice treated by EVs-miR-135a-5p mimic was higher, the cerebral infarction volume (Fig. 5H) and the concentration of IL-18 and IL-1β (Fig. 5I) in the brain supernatant were remarkably lower, while opposite effects were seen in mice treated by EVs-miR-135a-5p mimic + TXNIP compared with mice treated by EVs-miR-135a-5p mimic + NC mimic (Fig. 5G–I). In summary, M2 microglia-derived EVs deliver miR-135a-5p to inhibit TXNIP, thereby inhibiting the activation of NLRP3 inflammasome, reducing autophagic activity of neurons, and reducing ischemic brain injury.
Discussion
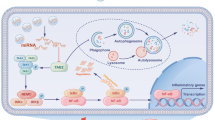
Irreversible injury and subsequent cell death in brain are induced by ischemia as occurs after diminished tissue blood flow leading to an imbalance of oxygen supply and energy demand [23, 24]. Ischemic brain injury can occur in infants as well as adults, caused a striking morbidity and mortality [25, 26]. Novel therapeutic agents for ischemia management are urgently demanded since existing agents are applicable for only a small portion of patients and may trigger adverse effects (e.g. tissue damage) [27]. A recent study has reported that EVs may play an important role in the development of brain ischemia [28]. Hence, this study aimed to investigate the function and molecular mechanism involving EVs derived from M2 microglial cells in ischemic brain injury. Our results illustrated that M2 microglia-derived EVs can inhibit TXNIP-mediated NLRP3 inflammasome activation through miR-135a-5p, thereby reduce neuronal autophagy and further alleviate ischemic brain injury in a mouse tMCAO model.
Primarily, this study identified that EVs derived from M2 microglia elevated the expression of p62 while reducing the expression of Beclin-1, LC3A/B, and LAMP-1 in the OGD/R cell model, and decreased the expression of IL-18 and IL-1β in the tMCAO mouse model, supporting that EVs derived from M2 microglia could inhibit neuronal autophagy and alleviate ischemic brain injury. Microglia are crucial cells that play immune functions in the central nervous system, and which are proposed as key players in the pathogenesis of neurodegenerative diseases and stroke injury [29]. Under ischemic conditions, microglia exhibit two opposite activation states or phenotypes: proinflammatory M1 and anti-inflammatory M2 phenotypes [30]. Therefore, the prospect of forcing microglial differentiation into the M2 polarized form to exert neuroprotective effects has received great attention, and studies have provided a theoretical basis for this approach and confirmed its feasibility [31, 32]. Moreover, EVs derived from microglial were identified as alleviators of neuroinflammatory diseases [8]. In addition, astrocyte-derived exosomes also repressed autophagy and ameliorated neuronal injury in ischemic stroke [33]. p62 is a multivalent protein that participates in cargo isolation in the process of selective autophagy, which was observed previously to have increased expression after therapeutic treatment of the OGD/R cell model [34, 35]. Beclin-1, LC3A/B, and LAMP-1 are all essential autophagic regulators, which have upregulated expression in the context of autophagy [36,37,38]. IL-18 and IL-1β are inflammation-related proteins, which show up-regulation after effective management in cerebral ischemia-reperfusion injury in rats [39]. Thus, EVs derived from M2 microglia serve as neuroprotective factors in ischemic brain injury.
Subsequently, we further uncovered that M2 microglia-derived EVs relieved ischemic brain injury by decreasing TXNIP-mediated NLRP3 inflammasome expression through miR-135a-5p. Indeed, previous work showed that MiR-135a protected against OGD/R-induced neuron injury [22]. Accumulating evidence indicates that EVs delivering miRNAs are involved in several pathological conditions, including ischemia [11, 40]. Furthermore, a recent study validated that M2 microglia-derived exosomes attenuated ischemic brain injury and promoted neuronal survival via exosomal miR-124, representing a promising avenue for treating ischemic stroke [21]. TXNIP is an enhancer of the internalization of glucose transporters, which is responsible for inflammasome activation and autophagy [41]. Downregulation of TXNIP was also verified to protect against cerebral ischemia and reperfusion injury by suppressing NLRP3 inflammasome activation [42]. Other research implicated NLRP3 in inflammatory response activation via activation and secretion of IL-1β and IL-18 [43]. Meanwhile, NLRP3 inflammasome inactivation was identified as a potential therapeutic target in ischaemic stroke [44]. Importantly, NLRP3 inflammasome activation was inhibited by miR-148a in M2 macrophage-derived exosomes through TXNIP inhibition, thereby combating against myocardial ischemia/reperfusion injury [45]. In summary, M2 microglia-derived EVs are the protective against ischemic injury through TXNIP-mediated alteration in NLRP3 inflammasome expression via miR-135a-5p.
Taken together, this study supported the proposal that an elevation of miR-135a-5p expression in M2 microglia-derived EVs inactivated the NLRP3 inflammasome through TXNIP downregulation, and repressed neuronal autophagy, all of which ultimately alleviated ischemic brain injury (Fig. 6). These early results in experimental models are promising for translational studies in the clinical setting testing the neuroprotective effect of M2 microglia-derived EVs.
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
References
Luheshi NM, Kovacs KJ, Lopez-Castejon G, Brough D, Denes A. Interleukin-1alpha expression precedes IL-1beta after ischemic brain injury and is localised to areas of focal neuronal loss and penumbral tissues. J Neuroinflammation. 2011;8:186.
He J, Gao Y, Wu G, Lei X, Zhang Y, Pan W, et al. Bioinformatics analysis of microarray data to reveal the pathogenesis of brain ischemia. Mol Med Rep. 2018;18:333–41.
Cabral-Miranda F, Nicoloso-Simoes E, Adao-Novaes J, Chiodo V, Hauswirth WW, Linden R, et al. rAAV8-733-mediated gene transfer of CHIP/Stub-1 prevents hippocampal neuronal death in experimental brain ischemia. Mol Ther. 2017;25:392–400.
Rahimian R, Cordeau P Jr, Kriz J. Brain response to injuries: when microglia go sexist. Neuroscience. 2019;405:14–23.
Jin WN, Shi SX, Li Z, Li M, Wood K, Gonzales RJ, et al. Depletion of microglia exacerbates postischemic inflammation and brain injury. J Cereb Blood Flow Metab. 2017;37:2224–36.
Katsuda T, Kosaka N, Takeshita F, Ochiya T. The therapeutic potential of mesenchymal stem cell-derived extracellular vesicles. Proteomics. 2013;13:1637–53.
Paolicelli RC, Bergamini G, Rajendran L. Cell-to-cell communication by extracellular vesicles: focus on microglia. Neuroscience. 2019;405:148–57.
Yang Y, Boza-Serrano A, Dunning CJR, Clausen BH, Lambertsen KL, Deierborg T. Inflammation leads to distinct populations of extracellular vesicles from microglia. J Neuroinflammation. 2018;15:168.
Zhu R, Liu X, Zhu Y, He Z. MiRNAs: potential diagnostic and therapeutic targets for cerebral ischaemia. Neurol Res. 2016;38:86–92.
Chen H, Li X. LncRNA ROR is involved in cerebral hypoxia/reoxygenation-induced injury in PC12 cells via regulating miR-135a-5p/ROCK1/2. Am J Transl Res. 2019;11:6145–58.
Yoshikawa FSY, Teixeira FME, Sato MN, Oliveira L. Delivery of microRNAs by extracellular vesicles in viral infections: could the news be packaged? Cells. 2019;8:611. https://doi.org/10.3390/cells8060611.
Zhang L, Liu H, Xu K, Ling Z, Huang Y, Hu Q, et al. Hypoxia preconditioned renal tubular epithelial cell-derived extracellular vesicles alleviate renal ischaemia-reperfusion injury mediated by the HIF-1alpha/Rab22 pathway and potentially affected by microRNAs. Int J Biol Sci. 2019;15:1161–76.
Chen XD, Zhao J, Yan Z, Zhou BW, Huang WF, Liu WF, et al. Isolation of extracellular vesicles from intestinal tissue in a mouse model of intestinal ischemia/reperfusion injury. Biotechniques. 2020;68:257–62.
Bharti V, Tan H, Chow D, Wang Y, Nagakannan P, Eftekharpour E, et al. Glucocorticoid upregulates thioredoxin-interacting protein in cultured neuronal cells. Neuroscience. 2018;384:375–83.
Zhou H, Tan H, Letourneau L, Wang JF. Increased thioredoxin-interacting protein in brain of mice exposed to chronic stress. Prog Neuropsychopharmacol Biol Psychiatry. 2019;88:320–6.
Kim GS, Jung JE, Narasimhan P, Sakata H, Chan PH. Induction of thioredoxin-interacting protein is mediated by oxidative stress, calcium, and glucose after brain injury in mice. Neurobiol Dis. 2012;46:440–9.
Yin Y, Zhou Z, Liu W, Chang Q, Sun G, Dai Y. Vascular endothelial cells senescence is associated with NOD-like receptor family pyrin domain-containing 3 (NLRP3) inflammasome activation via reactive oxygen species (ROS)/thioredoxin-interacting protein (TXNIP) pathway. Int J Biochem Cell Biol. 2017;84:22–34.
Huang J, Wang T, Yu D, Fang X, Fan H, Liu Q, et al. l-Homocarnosine attenuates inflammation in cerebral ischemia-reperfusion injury through inhibition of nod-like receptor protein 3 inflammasome. Int J Biol Macromol. 2018;118:357–64.
Guo ZN, Xu L, Hu Q, Matei N, Yang P, Tong LS, et al. Hyperbaric oxygen preconditioning attenuates hemorrhagic transformation through reactive oxygen species/thioredoxin-interacting protein/nod-like receptor protein 3 pathway in hyperglycemic middle cerebral artery occlusion rats. Crit Care Med. 2016;44:e403–11.
Chen D, Dixon BJ, Doycheva DM, Li B, Zhang Y, Hu Q, et al. IRE1alpha inhibition decreased TXNIP/NLRP3 inflammasome activation through miR-17-5p after neonatal hypoxic-ischemic brain injury in rats. J Neuroinflammation. 2018;15:32.
Song Y, Li Z, He T, Qu M, Jiang L, Li W, et al. M2 microglia-derived exosomes protect the mouse brain from ischemia-reperfusion injury via exosomal miR-124. Theranostics. 2019;9:2910–23.
Liu X, Li M, Hou M, Huang W, Song J. MicroRNA-135a alleviates oxygen-glucose deprivation and reoxygenation-induced injury in neurons through regulation of GSK-3beta/Nrf2 signaling. J Biochem Mol Toxicol. 2018;e22159. https://doi.org/10.1002/jbt.22159.
Radak D, Katsiki N, Resanovic I, Jovanovic A, Sudar-Milovanovic E, Zafirovic S, et al. Apoptosis and Acute Brain Ischemia in Ischemic Stroke. Curr Vasc Pharmacol. 2017;15:115–22.
Davidson JO, Green CR, Bennet L, Gunn AJ. Battle of the hemichannels–Connexins and Pannexins in ischemic brain injury. Int J Dev Neurosci. 2015;45:66–74.
Huang L, Zhang L. Neural stem cell therapies and hypoxic-ischemic brain injury. Prog Neurobiol. 2019;173:1–17.
Milanizadeh S, Zuwarali KNN, Aliaghaei A, Bigdeli MR. Therapeutic Potential of Pretreatment with Allograft Sertoli Cells Transplantation in Brain Ischemia by Improving Oxidative Defenses. J Mol Neurosci. 2018;64:533–42.
Yu-Hang L, Lun-De L, Su Jing C, Bandla A, Thakor NV. An integrated neuroprotective intervention for brain ischemia validated by ECoG-fPAM. Conf Proc IEEE Eng Med Biol Soc. 2016;2016:4009–12.
Zheng X, Bahr M, Doeppner TR. From tumor metastasis towards cerebral ischemia-extracellular vesicles as a general concept of intercellular communication processes. Int J Mol Sci. 2019;20:5995. https://doi.org/10.3390/ijms20235995.
Yang X, Xu S, Qian Y, Xiao Q. Resveratrol regulates microglia M1/M2 polarization via PGC-1alpha in conditions of neuroinflammatory injury. Brain Behav Immun. 2017;64:162–72.
Yu T, Yu H, Zhang B, Wang D, Li B, Zhu J, et al. Promising neuroprotective function for M2 microglia in kainic acid-induced neurotoxicity via the down-regulation of NF-kappaB and caspase 3 signaling pathways. Neuroscience. 2019;406:86–96.
Wang J, Xing H, Wan L, Jiang X, Wang C, Wu Y. Treatment targets for M2 microglia polarization in ischemic stroke. Biomed Pharmacother. 2018;105:518–25.
Xu S, Zhu W, Shao M, Zhang F, Guo J, Xu H, et al. Ecto-5’-nucleotidase (CD73) attenuates inflammation after spinal cord injury by promoting macrophages/microglia M2 polarization in mice. J Neuroinflammation. 2018;15:155.
Pei X, Li Y, Zhu L, Zhou Z. Astrocyte-derived exosomes suppress autophagy and ameliorate neuronal damage in experimental ischemic stroke. Exp Cell Res. 2019;382:111474.
Sanchez-Martin P, Sou YS, Kageyama S, Koike M, Waguri S, Komatsu M. NBR1-mediated p62-liquid droplets enhance the Keap1-Nrf2 system. EMBO Rep. 2020;21:e48902.
Wang Y, Ren Q, Zhang X, Lu H, Chen J. Neuroprotective mechanisms of calycosin against focal cerebral ischemia and reperfusion injury in rats. Cell Physiol Biochem. 2018;45:537–46.
Cai J, Zou Z, Wei S, Zheng Q, Xu Y, Lu Y, et al. Identification of Beclin-1 from orange-spotted grouper (Epinephelus coioides) involved in viral infection. Fish Shellfish Immunol. 2019;94:336–45.
Terasawa K, Tomabechi Y, Ikeda M, Ehara H, Kukimoto-Niino M, Wakiyama M, et al. Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes. Biochem Biophys Res Commun. 2016;479:489–95.
Lan R, Wu JT, Wu T, Ma YZ, Wang BQ, Zheng HZ, et al. Mitophagy is activated in brain damage induced by cerebral ischemia and reperfusion via the PINK1/Parkin/p62 signalling pathway. Brain Res Bull. 2018;142:63–77.
Meng C, Zhang J, Zhang L, Wang Y, Li Z, Zhao J. Effects of NLRP6 in cerebral ischemia/reperfusion (i/r) injury in rats. J Mol Neurosci. 2019;69:411–8.
Gu H, Liu Z, Li Y, Xie Y, Yao J, Zhu Y, et al. Serum-derived extracellular vesicles protect against acute myocardial infarction by regulating miR-21/PDCD4 signaling pathway. Front Physiol. 2018;9:348.
Dafre AL, Schmitz AE, Maher P. Rapid and persistent loss of TXNIP in HT22 neuronal cells under carbonyl and hyperosmotic stress. Neurochem Int. 2020;132:104585.
Hou Y, Wang Y, He Q, Li L, Xie H, Zhao Y, et al. Nrf2 inhibits NLRP3 inflammasome activation through regulating Trx1/TXNIP complex in cerebral ischemia reperfusion injury. Behav Brain Res. 2018;336:32–9.
Nakata A, Nadatani Y, Watanabe T, Matsumoto Y, Kosaka S, Higashimori A, et al. NOD-like receptor family pyrin domain-containing 3 inflammasome activation exacerbates 5-fluorouracil-induced small intestinal mucositis via interleukin-1beta activation. Digestion. 2021;102:298–312. https://doi.org/10.1159/000504974.
Ito M, Shichita T, Okada M, Komine R, Noguchi Y, Yoshimura A, et al. Bruton’s tyrosine kinase is essential for NLRP3 inflammasome activation and contributes to ischaemic brain injury. Nat Commun. 2015;6:7360.
Dai Y, Wang S, Chang S, Ren D, Shali S, Li C, et al. M2 macrophage-derived exosomes carry microRNA-148a to alleviate myocardial ischemia/reperfusion injury via inhibiting TXNIP and the TLR4/NF-kappaB/NLRP3 inflammasome signaling pathway. J Mol Cell Cardiol. 2020;142:65–79.
Author contributions
YL, Y-PL, and C-HX conceived and designed research. YL, Y-PL, and L-MX performed experiments. L-KC and S-YZ, analyzed data. YL, Y-PL, L-MX, and C-HX interpreted results of experiments. E-MZ prepared figures. L-KC, S-YZ, E-MZ, drafted manuscript. YL, Y-PL, L-MX, and C-HX edited and revised manuscript. All authors approved final version of manuscript.
Funding
The study was supported by Natural Science Foundation of Jiangxi Province (No.20192BAB205067).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
The current study was performed with the approval of the Ethics Commission of The First Affiliated Hospital of Nanchang University. All animal experiments were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and relevant national regulations.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Y., Li, YP., Xiao, LM. et al. Extracellular vesicles derived from M2 microglia reduce ischemic brain injury through microRNA-135a-5p/TXNIP/NLRP3 axis. Lab Invest 101, 837–850 (2021). https://doi.org/10.1038/s41374-021-00545-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-021-00545-1
This article is cited by
-
The Potential of NLRP3 Inflammasome as a Therapeutic Target in Neurological Diseases
Molecular Neurobiology (2023)
-
Up-regulating microRNA-214-3p relieves hypoxic–ischemic brain damage through inhibiting TXNIP expression
Molecular and Cellular Biochemistry (2023)
-
Molecular Mechanism of the Protective Effects of M2 Microglia on Neurons: A Review Focused on Exosomes and Secretory Proteins
Neurochemical Research (2022)